
Pros and cons of different ways to address dysfunctional autophagy in Pompe disease
Introduction
A large number of lysosomal storage disorders (LSDs), including Pompe disease, underscore the importance of the lysosome—a ubiquitous intracellular organelle enclosed in a membrane that maintains and protects the acidic interior. The scientific discoveries over the past decade have fundamentally changed our understanding of the role of the lysosome in cellular metabolism—from the view of this organelle as a terminal waste bag to a sophisticated “switchboard” center involved in arranging the cellular response to various metabolites and multiple types of stress and maintaining a balance between anabolic and catabolic processes, including autophagy.
Autophagy (from Greek “self-eating”) is an evolutionarily conserved lysosome-dependent recycling process that provides nutrients at times of starvation and clears the cell of protein aggregates and worn-out or dysfunctional intracellular organelles. At least three forms of autophagy are recognized: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy (1). In microautophagy, cytosolic cargos are “swallowed” by the lysosome through the inward folding of the lysosomal membrane itself (2). CMA refers to a selective degradation of a subset of soluble cytosolic proteins containing a pentapeptide motif (KFERQ-like). This motif serves as a tag for the recognition by a chaperone (the heat shock protein Hsc70) that delivers the protein to the lysosomal surface; once there, the substrate protein translocates across the lysosomal membrane via the lysosome-associated membrane protein type 2A (LAMP2A)-mediated mechanism [reviewed in (3)]. Macroautophagy is by far the most studied form, and it is unique in that it relies on the de novo formation of a transient double-membrane vesicle, called autophagosome, that mediates the delivery of cytosolic cargo to lysosomes. In this review we focus on the basics of macroautophagy (subsequently referred to as autophagy as is commonly accepted in the literature) and its relevance to the pathophysiology of Pompe disease. We will also discuss different ways to manipulate the autophagic process to provide a therapeutic benefit for this severe muscle disorder.
Brief overview of autophagy
Autophagy was initially defined as a process that allows the cell to survive under starvation conditions by digesting its own components so that the degradation products can be exported back to the cytoplasm and used for protein synthesis and energy production during “lean” times. In the course of this multi-step process, newly formed double-membrane cup-shaped structures, called phagophores, surround portions of the cytoplasm and become autophagosomes after their edges are sealed. Autophagosomes, in which cytosolic components are sequestered, move along microtubules and fuse with lysosomes, giving rise to autolysosomes where the contents of autophagosomes are degraded by lysosomal hydrolases [reviewed in (1,4,5)]. Once the degradation is completed, autolysosomes contribute to the regeneration of the lysosomal pool (6). Autophagosomes can also fuse with vesicles of the endocytic pathway—late endosomes—to form amphisomes which then fuse lysosomes (7). This “prelysosomal connections” between autophagy and endocytosis (8) may have implications for enzyme replacement therapy (ERT) for Pompe disease as well as other LSDs since the recombinant enzymes traffic to the lysosome along the endocytic pathway (9).
Unlike proteasome-dependent degradation of polyubiquitin-labeled proteins, autophagic response to starvation is a non-selective “wholesale” degradation of cytoplasmic materials which are randomly sequestered into autophagosomes. However, the system also operates at baseline levels even when nutrients are available; under this conditions autophagy fulfills housekeeping functions and contributes to intracellular homeostasis by selective removal of potentially harmful products, such as aberrant protein aggregates (that cannot by degraded by proteasomes), damaged or no longer functional organelles, and pathogens [reviewed in (10-12)].
Autophagy is a highly regulated process; uncontrolled autophagy may have serious repercussions for the cell. Dozens of proteins are responsible for multiple steps of the autophagic pathway—the initiation of autophagy, formation and maturation of autophagosomes, autophagosomal-lysosomal fusion, and cargo degradation. We refer the reader to a number of recent reviews covering the molecular machinery of autophagy (5,13,14). The two proteins that are most commonly used as autophagy markers in mammals are microtubule associated protein 1 light chain 3 (MAP1LC3; LC3) and sequestosome 1 (SQSTM1; also known as p62). LC3, a mammalian homologue of yeast ATG8, is a soluble protein that exists as a cytosolic LC3-I form and as a phosphatidylethanolamine-conjugated LC3-II form. The latter is recruited to both the inner and outer membranes of forming autophagosomes where it remains all the way throughout their fusion with lysosomes; LC3-II serves as a highly specific marker of autophagosomes (15). SQSTM1/p62 is a ubiquitin-binding scaffold protein with a variety of cellular functions. This protein plays a dual role in autophagy: it is primarily degraded by autophagy, thus, serving as an autophagy substrate, and it acts as a receptor in selective autophagy by interacting with LC3 [and gamma-aminobutyric acid receptor-associated protein (GABARAP family proteins)] to promote autophagic degradation of ubiquitinated protein aggregates (16-18). The protein accumulates in cells from autophagy-deficient mice (19), and an increase in the level of p62 is an indication of a functional defect of autophagy.
One must keep in mind that autophagy is a dynamic process which has a beginning—autophagosome formation, and an end—fusion with the lysosome and lysosomal digestion of the cargo. Autophagic flux refers to the overall efficiency of the process, namely, to the rate of autophagosome content degradation. The flux is complete if the formation of autophagosomes is followed by their fusion with lysosomes and lysosomal degradation of the cargos; if, however, the formation of autolysosomes is impaired/or lysosomal degradation is insufficient, the flux is incomplete, leading to accumulation of autophagosomes and autophagy substrates (1). Therefore, the increase in the number of autophagosomes could indicate an upregulation of autophagy or defects in autophagosome-lysosome fusion, or both.
As mentioned above, autophagy can be highly selective process, in which the cargo is labeled and recruited to the autophagosomal membrane by the receptors/or adaptors (10-12). Multiple pathways target specific cellular compartments for lysosomal degradation, and among these, the most studied is autophagic removal of damaged mitochondria—a process called mitophagy (20). A decrease in mitochondrial membrane potential activates PINK1 kinase followed by Parkin-mediated recruitment of autophagic machinery (mutations in both proteins are often found in patients with early-onset of Parkinson’s disease) (21) [reviewed in (22)]. It is not surprising that aberrant autophagic turnover of dysfunctional mitochondria has been implicated in several LSDs, including Pompe disease (23). Abnormal mitochondrial function and inefficient clearance of damaged mitochondria in muscle were documented in GAA-KO mice (24). Altered mitochondrial morphology is commonly observed in muscle biopsies from Pompe disease patients (25,26).
In addition to mitophagy, other kinds of selective autophagy have been identified: pexophagy for peroxisomes, ribophagy for ribosomes, endoplasmic reticulum (ER)-phagy for degradation of the ER, nucleophagy for selective targeting portion of the nucleus, lipophagy for selective degradation of lipid droplets, etc. Two additional specific forms of selective autophagy—lysophagy and glycophagy—will be discussed in greater detail because of their potential relevance to Pompe disease.
Lysophagy and glycophagy
Lysophagy, by definition, involves selective elimination of damaged lysosomes through the autophagic process. Lysosome damage, called lysosomal membrane permeabilization (LMP), and lysosomal rupture are prominent features of skeletal muscle damage in Pompe disease (25). In the course of examining muscle fibers (immunostained for lysosomal and autophagosomal markers) isolated from biopsies of Pompe disease patients and GAA-KO mice, we frequently observed irregularly-shaped lysosomes inside autophagosomes. Based on this morphological evidence, we suggested that damaged lysosomes may trigger an increase in autophagy and proposed the term lysophagy long before the molecular mechanism of this type of selective autophagy began to unravel (27).
In an elegant study, using the light-based lysosome-inactivation technique to injure only a subset of lysosomes in a cell, Hung et al. demonstrated selective ubiquitination and recruitment of damaged lysosomes by autophagic proteins followed by their incorporation into autolysosomes (28). Induction of autophagy and selective engulfment of damaged lysosomes by autophagosomes have been also shown in the murine cells treated with lysosomotropic agents (29). The lysosome-containing autophagosomes are then likely to fuse with intact lysosomes leading to the restoration of proteolytic activity. The relevance of lysophagy and its protective role was demonstrated in vivo, in a mouse model of acute hyperuricemic nephropathy (29). Lysophagy may also play a protective role in Danon disease, caused by X-linked mutations in LAMP2a gene encoding a lysosomal membrane protein. It has been suggested that the recruitment of LGALS3 (galectin-3; a marker of endovascular damage) to the damaged lysosomal membrane may be one of the triggers of lysophagy (30). Although the mechanisms of selective degradation of lysosomes are not fully understood, recent studies indicate that cytosolic galectins induce autophagy in response to lysosomal damage by controlling mTOR and AMPK (5' AMP-activated protein kinase) (31). The contribution of the lysophagy to the pathogenesis of muscle damage in Pompe disease is a focus of our current investigation at the NIH.
Glycophagy is a somewhat misleading term. Strictly speaking, it implies that abnormal glycogen is selectively targeted to the lysosome for degradation. Although aberrant glycogen structure/or abnormal branching has been considered as possible “labels” for selective shunting through autophagic degradation (32), there is no solid data to support the hypothesis. Similarly, there is no data showing that accumulated lysosomal glycogen in Pompe disease is abnormal.
In general, despite extensive research into mechanisms involved in the pathogenesis of Pompe disease, the most obvious question of how (and why) glycogen gets to the lysosome in the first place is still not fully addressed. Early reports provided morphological evidence of autophagic delivery of glycogen to the lysosomes in skeletal muscle of neonatal rats; the presence of large glycogen-filled autophagic vacuoles in muscle of newborn animals suggested the role of lysosome in glycogen breakdown at birth, when the demand for glucose is high (33). The appearance and timing of these vacuoles (containing almost exclusively glycogen particles), their absence in fetal tissues, and their decline in number in the first few days after birth, all suggested a selective nature of the physiological process of postnatal glycogen mobilization (33). More recent studies indicated that selective and highly regulated (by the cyclic AMP and the mTOR pathway) glycogen autophagy also takes place in the liver and heart to meet the demand for glucose in the postnatal period (34-37).
Our studies pointed to the role of glycogen autophagy in skeletal muscle of adult animals: genetic suppression of autophagy by inactivation of a critical autophagic gene, Atg7, in muscles of GAA-KO mice (Atg7/GAA double knockout) significantly reduced the amount of lysosomal glycogen accumulation. This suggested that autophagic pathway is, at least partially, responsible for glycogen trafficking to the lysosome in adults (38).
Autophagic delivery of glycogen particles to the lysosome could simply reflect their cytoplasmic distribution. On the other hand, it has been suggested that glycogen can be selectively transported to the lysosome through the autophagic pathway. The starch-binding domain-containing protein 1 (STBD1; genethonin 1) which contains the carbohydrate-binding domain, was proposed to function as a novel receptor for anchoring glycogen to the autophagosomal membrane through interaction with the cognate autophagy protein GABARAPL1—a process termed “glycophagy” (39). If, indeed, STBD1 is a mediator of glycogen trafficking to the lysosome, then its inhibition would rescue or ameliorate glycogen burden in Pompe disease. However, adeno-associated virus (AAV)-mediated inhibition of STBD1 in GAA-KO mice did not change the levels of lysosomal glycogen accumulation in the diseased muscle (40). Furthermore, in GAA/STBD1 double knockouts, glycogen accumulation was significantly reduced in the liver but not in skeletal and cardiac muscles, suggesting a tissue-specific role of Stbd1 in glycogen transport (41).
Recent studies in Drosophila melanogaster raise an interesting possibility that glycogen synthase (GlyS), a protein long-known to be involved in glycogen synthesis, regulates autophagy through its interaction with Atg8 (42). To study glycogen autophagy in skeletal muscle, Zirin et al. (42) established an in vivo fruit fly model of vacuolar myopathy by using chloroquine (CQ), a widely used antimalarial drug. Prolonged CQ treatment is associated with the development of a vacuolar myopathy (43) due to its lysosomotropic effect and disruption of autophagosomal-lysosomal fusion (44). Of note, early clinical case reports indicate that glycogen is a major component of autophagic vacuoles in CQ myopathy (45,46).
Nutrient deprivation in CQ-treated fruit flies triggered massive glycogen autophagy in the larval muscle as indicated by: (I) the high degree of colocalization of glycogen and an autophagic marker; and (II) the presence of numerous autophagic vesicles loaded with glycogen on electron micrographs. Knockdown of GlyS reduced the size but not the number of autophagosomes in starved CQ-treated muscles suggesting an important role of GlyS in CQ-induced autophagosome swelling. These data, combined with the evidence of the interaction between GlyS and Atg8, led to a hypothesis that GlyS on the surface of the autolysosome may sense glucose released by lysosomal glycogen degradation (42). Furthermore, genetic screen using primary myocyte cells from a fruit fly model of autophagy confirmed a critical role of glycogen metabolites in modulating autophagic response to starvation in muscle (47). Glycogen autophagy has also been suggested as a component of the response to cardiac metabolic stress in female hearts (48).
From our perspective, the role of glycophagy in the pathogenesis of muscle damage in Pompe disease is, no doubt, a high-priority research area.
Dysfunctional autophagy in Pompe disease
One of the earliest description of patients’ muscle biopsies (long before the field of autophagy began to advance), included “vacuolar myopathy, high vacuolar glycogen content, and autophagic character of many vacuoles”. The described pathology was reminiscent to that observed in human and experimentally induced CQ myopathy (49)—a profound insight that makes perfect sense now.
In hindsight, predicting the involvement of autophagy in Pompe disease, should have been a slam-dunk. But, as often happens, the new concept of the pathogenesis of muscle damage took a long time to develop. For years, a model of disease progression, broken down into stages of lysosomal enlargement and rupture was considered sufficient (25,50). To be more specific, autophagy emerged as a player when the shortcomings of ERT came to light. Efforts to advance the development of ERT for Pompe disease in the 1990s have been rewarded with the approval of in 2006 of human recombinant GAA (rhGAA, alglucosidase alfa; Myozyme®, Sanofi Genzyme, Cambridge, MA, USA). Although initial reports had evoked optimism, our experience with rhGAA in GAA-KO mice was not encouraging.
While the response of cardiac muscle to ERT was very good, skeletal muscle improvement remained far less satisfactory. Most of the enzyme ended up in the liver, large doses (20–40 mg/kg/wk)—far exceeding those used in other LSDs, failed to clear skeletal muscle glycogen. Even with huge doses (100 mg/kg), considerable amount of glycogen remained in skeletal muscle (51). When company-sponsored human clinical trials were completed, the results resembled those in our pre-clinical studies. Skeletal muscle has proved a recalcitrant target. Over the years it became increasingly clear that sub-optimal delivery of the drug to skeletal muscle as well as the condition within muscle cells themselves contribute to the poor muscle response to therapy.
Electron microscopy (EM) of skeletal muscle biopsies from Pompe disease patients (particularly with late-onset disease) and from GAA-KO mice showed large areas of autophagic accumulation containing classical double-membrane autophagosomes with undigested materials or glycogen particles, multivesicular bodies, and multimembrane structures. Immunostaining of single muscle fibers for lysosomal (LAMP1) and autophagosomal (LC3) markers reveals the extent of autophagic buildup: the core of muscle fibers was packed with clusters of late endosomes/lysosomes with broken borders, autophagosomes, autofluorescent material, and other cellular debris. The extensive autophagic buildup in the core of muscle fibers, rather than lysosomal expansion, seemed to disrupt sarcomere structure leading to the loss of muscle force in GAA-KO mice (52). Likewise, autophagic buildup was the overwhelming pathology in muscle biopsies from children and adults with Pompe disease. In addition, the buildup area was filled with undigested autophagic substrates, such as p62/SQSTM1 and potentially toxic ubiquitinated protein aggregates, suggesting the failure of the autophagic process in the diseased muscle (27,53,54). Gene expression analysis, western blotting, and live-cell imaging indicated that both, increased formation of autophagosomes and impaired autophagosomal-lysosomal fusion (autophagic block; Figure 1) in muscle cells underlie the mechanism of autophagic accumulation (55,56). Nowadays, Pompe disease is classified as autophagic myopathy (57), and dysregulation of autophagy is recognized as a common feature of many LSDs (58,59).
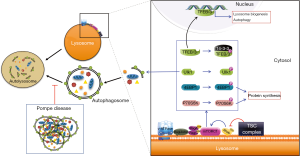
The consequences of autophagic defect in Pompe skeletal muscle are twofold: (I) massive autophagic buildup (non-contractile inclusions) within the fibers disrupts the muscle architecture by interrupting the contractile myofibrils and negatively affects muscle function by causing loss of muscle force and contractility (60-62); and (II) the buildup disturbs vesicular movement and presents an obstacle for the trafficking and lysosomal delivery of the therapeutic enzyme (56,63-65). Indeed, in preclinical trials, poor skeletal muscle response to ERT was linked to the presence of autophagic buildup (55,63). Several strategies have been explored to address the dysfunctional autophagy in GAA-KO mice.
Stimulation of autophagosomal-lysosomal fusion and lysosomal exocytosis
One way to overcome the autophagic block is to restore the fusion between autophagosomes and lysosomes. The feasibility of this approach came with the discovery of the role of transcription factors, TFEB and TFE3, in the biogenesis of lysosomes and autophagosomes. When translocated to the nuclei, both TFEB and TFE3 stimulate the generation of new lysosomes and autophagosomes and promote fusion between them (66,67). The mechanism of TFEB/TFE3-mediated gene regulation involves their ability to bind a 10-base pair palindromic sequence (called CLEAR: Coordinated Lysosomal Expression and Regulation) in the promoter regions of many lysosomal and autophagic genes (66,67). The TFEB/TFE3 cytoplasmic-nuclear shuttling is controlled by the mammalian target of rapamycin (mTORC1). When nutrients are available, TFEB and TFE3 are recruited to the lysosomal membranes where they are phosphorylated by mTOR (at S211 of TFEB and S321 of TFE3), creating a binding site for the chaperone 14-3-3, thus keeping them in the cytosol; upon nutrient deprivation, mTORC1 is inactivated and dephosphorylated TFEB and TFE3 translocate to the nucleus to induce lysosomal biogenesis and autophagy (66-70).
In addition to regulating the transcription of genes involved autophagosomal-lysosomal biogenesis, TFEB and TFE3 also facilitate lysosomal exocytosis—a process of lysosomal fusion with plasma membrane leading to clearance of pathologically accumulated materials by discharging lysosomal content into the extracellular space (71). The ability of lysosomes to exocytose is now seen as an essential part of their function, namely, to promote cellular clearance by degrading the cargo or by releasing it outside the cell (68,72,73).
Overexpression of TFEB emerged as a potential therapeutic strategy in a variety of lysosomal storage disease as well as in disorders with accumulation of abnormal proteins (66,71). Indeed, overexpression of TFEB and TFE3 in GAA-deficient cultured muscle cells and in a particular muscle group (flexor digitorum brevis, FDB) of the GAA-KO mice triggered lysosomal exocytosis, reduced glycogen load and lysosomal size, and alleviated autophagic buildup (56,67,74). Also, overexpression of TFEB combined with GAA gene transfer promoted glycogen clearance and improved muscle pathology in iPSC-derived skeletal muscle cells from Pompe disease patients (75). Stimulation of lysosomal exocytosis—a conceptually novel approach to therapy, which addressed both lysosomal and autophagic pathologies in the diseased muscle, seemed very promising for treatment of Pompe disease.
However, the results of AAV-mediated systemic delivery of the human TFEB gene [under the control of the muscle creatine kinase (MCK) promoter] into GAA-KO mice were far less impressive, to put it mildly: TFEB treatment did not cause a significant decrease in total glycogen levels in all tissues examined, including skeletal muscles, diaphragm, or heart (76). It turned out that in skeletal muscle (unlike in other tissues) both TFEB and TFE3 stimulate expression of genes involved in several pathways related to glucose homeostasis and mitochondrial biogenesis. Increase in glucose uptake and glycogen synthesis was observed in transgenic mice expressing Tfe3 in skeletal muscle (77). A metabolic effect leading to increased muscle glycogen was also observed in muscle-specific TFEB transgenic mice (78).
Although disappointing, a discussion about the merits of this approach should continue. One can envisage a possibility of pharmacological modulation of endogenous TFEB/TFE3 in combination with ERT. An important caveat is that inhibition of mTORC1 to stimulate lysosomal exocytosis (for example, by rapamycin) should be avoided considering that the activity of this kinase is already reduced in the diseased muscle (see below).
Genetic suppression of autophagy in skeletal muscle
The association between defective autophagy and muscle resistance to ERT suggested that the removal of autophagic buildup would be helpful. The generation of autophagy deficient GAA-KO mice (Atg7/GAA double knockout) by genetic suppression of autophagy in skeletal muscle served this purpose. As mentioned above, a significant decrease in the amount of accumulated muscle glycogen was observed in double knockouts, indicating that this approach served as a substrate reduction therapy. However, this genetic manipulation did not completely eliminate lysosomal glycogen load, suggesting that in addition to autophagy, other pathways are involved in glycogen trafficking to the lysosome, such as, for example, microautophagy (38).
Perhaps, most important, without autophagic buildup, ERT worked very well in both young (3 mo. old) and old (7–8 mo. old) animals, as shown by complete or near complete clearance of stored glycogen and reversal of lysosomal pathology, an outcome never observed in GAA-KO with genetically intact autophagy (38,79).
The loss of autophagy in skeletal muscle of healthy mice is associated with the accumulations of dysfunctional mitochondria, mild atrophy, and age-dependent decrease in muscle strength (80-82). On the other hand, in the context of GAA deficiency, the benefits outweigh the negatives. Atg7/GAA double knockouts are phenotypically indistinguishable from the GAA-KO mice, and both strains have normal lifespans. Furthermore, the force level generated by single fibers from double knockouts was significantly higher than in GAA-KO, although not as high as in the wild type mice (79).
Thus, the removal of autophagic buildup in muscle of GAA-KO mice resulted in a decrease in glycogen level, increase in muscle force, and rendered skeletal muscle fully amenable to ERT. On the flip side, this approach is, indeed, leads to accumulation of SQSTM1/p62 and ubiquitinated proteins, and exacerbates ER stress and muscle atrophy. These experiments are proof of principle designed to explore the pathway of the cytosol-to-lysosome glycogen transport and to support the idea that autophagic buildup represents an impediment to ERT. An additional factor likely to contribute to the success of ERT in double knockouts is a significantly reduced muscle glycogen load.
Modulation of mTOR signaling
Since the nature of autophagy impairment in Pompe skeletal muscle involves both induction of autophagy and faulty autophagosomal-lysosomal fusion, most viable strategy may be inhibition of autophagosome formation. mTORC1 kinase, a nutrient sensor, along with AMPK, an intracellular sensor of ATP, respond to changing metabolic and energy conditions to coordinate anabolic (mRNA translation) and catabolic pathways (such as autophagy) (83-85). mTORC1 and AMPK can directly regulate autophagy with opposite effects by coordinated phosphorylation of autophagy-initiating kinase Ulk1: mTORC1 negatively regulates Ulk1 by the inactivating phosphorylation at Ser757 leading to a robust suppression of autophagosome formation (Figure 1), whereas AMPK catalyzes activating phosphorylation of Ulk1 at different sites (Ser317 and S777) leading to autophagy induction (86). When nutrients are abundant, active mTORC1 inhibits autophagy and prevents AMPK-mediated phosphorylation of Ulk1. When the cell experiences energy deficit or glucose starvation, AMPK is activated, and autophagy is induced.
Apart from the control of autophagy, both mTORC1 and AMPK regulate protein translation. When activated, mTORC1 stimulates protein synthesis by phosphorylating several downstream targets including the two well-established main regulators of cap-dependent protein synthesis—p70 ribosomal S6 kinase (P70S6K) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4EBP1) (Figure 1). AMPK inhibits mTORC1 through multiple mechanisms including activating phosphorylation of the tuberous sclerosis complex (TSC) which inhibits Rheb (Ras homolog enriched in brain)—a powerful mTOR activator (87). Recent studies demonstrated that the lysosomal membrane is the major site for mTORC1 activation. Multiple proteins including v-ATPase, Ragulator, Rag and Rheb GTPases, TSC complex, and others are engaged in the recruitment of mTORC1 to the lysosome and its release from the lysosome (inactivation) (Figure 1). Under nutrient-rich conditions, mTORC1 is recruited to the lysosome; conversely, amino-acid starvation triggers mTORC1 lysosomal release and inactivation (87-92). This breakthrough discovery combined with the established role of mTOR in controlling muscle mass (93) underscores the therapeutic potential of manipulating mTORC1 activity in the diseased muscle.
A reduced insulin-stimulated mTORC1 activation was described in two cellular models of Pompe disease—GAA-deficient C2C12 myoblasts and human fibroblasts from infantile Pompe disease patients (94). The disturbance of mTORC1 signaling was also reported in a recently developed an in vitro model of infantile onset Pompe disease using patient-specific iPSCs differentiated into myocytes (95). We have done extensive analysis of the mTORC1 status in cultured GAA-deficient myotubes and in muscle of GAA-KO mice (96). The results of this latter study can be briefly summarized as follows: mTORC1 is unable to properly shuttle to and from the lysosome as demonstrated by immunostaining of myotubes with LAMP1/mTORC1 under nutrient-rich and starved conditions; the basal activity of mTORC1 is reduced as shown by a decrease in the phosphorylation levels of its two downstream targets; the excess of TSC at the lysosomal surface and activation of AMPK-TSC pathway are responsible for the diminished mTOR activity, which contributes to muscle wasting (96).
These data provided the basis for developing a new therapeutic approach—mTORC1-mediated inhibition of autophagy. Indeed, reactivation of dysregulated mTORC1 by knocking down TSC2 (the Rheb inhibitor) rescued autophagy defect and reversed muscle atrophy in GAA-KO mice. Furthermore, the aberrant mTOR signaling was restored by arginine supplementation (96). Importantly, this approach demonstrated the feasibility of reversing the fully established autophagic buildup. Once again, as in the case of genetic suppression of autophagy, the removal of autophagic buildup following AAV-mediated TSC knockdown in GAA-KO mice resulted in efficient cellular clearance on ERT (79). Thus, by bringing mTORC1 activity to “normal” levels, one can achieve several goals—alleviation of autophagic pathology, increase in muscle mass, and better response to ERT.
Next-generation of ERT
Finally, the question is whether a more efficient recombinant GAA with better muscle-targeting properties can alone alleviate autophagic pathology. Recent study by Xu et al. (97) suggests that the answer is “yes”. A direct comparison of the effect of the currently available drug and a newly developed rhGAA (ATB200; Amicus proprietary recombinant human acid alpha-glucosidase) in GAA-KO mice demonstrated the superiority of the next-generation of ERT. Unlike alglucosidase alfa, ATB200 has high levels of mannose 6-phosphate (M6P) that are required for efficient cellular uptake and lysosomal trafficking. The enzyme is administered in combination with the pharmacological chaperone AT2221 (miglustat), which improves its pharmacokinetic properties. ATB200/AT2221 efficiently reversed lysosomal glycogen accumulation in muscle cells and significantly reduced the number of fibers with autophagic buildup—a striking contrast compared to alglucosidase alfa. These data provide the first evidence that the buildup can be resolved by ERT. The efficient lysosomal glycogen clearance and the generation of a pool of healthy lysosomes capable of digesting autophagic debris may explain the finding. Another possibility is that on the way to lysosomes along the endocytic pathway, ATB200/AT2221 may degrade glycogen that is trapped in amphisomes/autolysosomes, the structures which are sufficiently acidified (98) to allow for the drug to exert its function. However, it is still unclear whether the long-term ATB200/AT2221 therapy would fully reverse the autophagic pathology and the success of therapy is likely to depend on the timing of intervention. Lastly, the possibility remains that an adjunct therapy, such as, for example, mTORC1-mediated inhibition of autophagy, will be required in addition to the next generation of ERT. Whatever the outcome, this novel drug brings renewed hope.
Acknowledgments
This research was supported by the Intramural Research Program of the NHLBI of the National Institutes of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931-7. [Crossref] [PubMed]
- Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cellular and molecular life sciences CMLS 2012;69:1125-36. [Crossref] [PubMed]
- Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 2018;19:365-81. [Crossref] [PubMed]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005;118:7-18. [Crossref] [PubMed]
- Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J 2017;36:1811-36. [Crossref] [PubMed]
- Yu L, McPhee CK, Zheng L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010;465:942-6. [Crossref] [PubMed]
- Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 1988;151:40-7. [Crossref] [PubMed]
- Gordon PB, Hoyvik H, Seglen PO. Prelysosomal and lysosomal connections between autophagy and endocytosis. Biochem J 1992;283:361-9. [Crossref] [PubMed]
- Neufeld EF. From serendipity to therapy. Annu Rev Biochem 2011;80:1-15. [Crossref] [PubMed]
- Hubbard VM, Valdor R, Macian F, et al. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology 2012;13:21-35. [Crossref] [PubMed]
- Jin M, Liu X, Klionsky DJ. SnapShot: Selective autophagy. Cell 2013;152:368-368.e2. [Crossref] [PubMed]
- Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 2018;20:233-42. [Crossref] [PubMed]
- Cooper KF. Till Death Do Us Part: The Marriage of Autophagy and Apoptosis. Oxid Med Cell Longev 2018;2018:4701275. [Crossref] [PubMed]
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 2018;19:349-64. [Crossref] [PubMed]
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720-8. [Crossref] [PubMed]
- Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005;171:603-14. [Crossref] [PubMed]
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131-45. [Crossref] [PubMed]
- Bjorkoy G, Lamark T, Pankiv S, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009;452:181-97. [Crossref] [PubMed]
- Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007;131:1149-63. [Crossref] [PubMed]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011;12:9-14. [Crossref] [PubMed]
- Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008;183:795-803. [Crossref] [PubMed]
- Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci 2012;125:795-9. [Crossref] [PubMed]
- Lim JA, Kakhlon O, Li L, et al. Pompe disease: Shared and unshared features of lysosomal storage disorders. Rare Dis 2015;3:e1068978. [Crossref] [PubMed]
- Lim JA, Li L, Kakhlon O, et al. Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy 2015;11:385-402. [Crossref] [PubMed]
- Thurberg BL, Lynch MC, Vaccaro C, et al. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for pompe disease. Lab Invest 2006;86:1208-20. [Crossref] [PubMed]
- Schoser BG, Muller-Hocker J, Horvath R, et al. Adult-onset glycogen storage disease type 2: clinico-pathological phenotype revisited. Neuropathol Appl Neurobiol 2007;33:544-59. [PubMed]
- Raben N, Takikita S, Pittis MG, et al. Deconstructing Pompe disease by analyzing single muscle fibers. Autophagy 2007;3:546-52. [Crossref] [PubMed]
- Hung YH, Chen LM, Yang JY, et al. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 2013;4:2111. [Crossref] [PubMed]
- Maejima I, Takahashi A, Omori H, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 2013;32:2336-47. [Crossref] [PubMed]
- Kawabata T, Yoshimori T. Beyond starvation: An update on the autophagic machinery and its functions. J Mol Cell Cardiol 2016;95:2-10. [Crossref] [PubMed]
- Jia J, Abudu YP, Claude-Taupin A, et al. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy 2019;15:169-71. [Crossref] [PubMed]
- Roach PJ, Depaoli-Roach AA, Hurley TD, et al. Glycogen and its metabolism: some new developments and old themes. Biochem J 2012;441:763-87. [Crossref] [PubMed]
- Schiaffino S, Hanzlikova V. Autophagic degradation of glycogen in skeletal muscles of the newborn rat. J Cell Biol 1972;52:41-51. [Crossref] [PubMed]
- Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc Res Tech 2004;64:10-20. [Crossref] [PubMed]
- Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract 2006;202:631-8. [Crossref] [PubMed]
- Kondomerkos DJ, Kalamidas SA, Kotoulas OB. An electron microscopic and biochemical study of the effects of glucagon on glycogen autophagy in the liver and heart of newborn rats. Microsc Res Tech 2004;63:87-93. [Crossref] [PubMed]
- Kondomerkos DJ, Kalamidas SA, Kotoulas OB, et al. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol 2005;20:689-96. [PubMed]
- Raben N, Schreiner C, Baum R, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder-murine Pompe disease. Autophagy 2010;6:1078-89. [Crossref] [PubMed]
- Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun 2011;413:420-5. [Crossref] [PubMed]
- Yi H, Fredrickson KB, Das S, et al. Stbd1 is highly elevated in skeletal muscle of Pompe disease mice but suppression of its expression does not affect lysosomal glycogen accumulation. Mol Genet Metab 2013;109:312-4. [Crossref] [PubMed]
- Sun T, Yi H, Yang C, et al. Starch Binding Domain-containing Protein 1 Plays a Dominant Role in Glycogen Transport to Lysosomes in Liver. J Biol Chem 2016;291:16479-84. [Crossref] [PubMed]
- Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 2013;11:e1001708. [Crossref] [PubMed]
- Casado E, Gratacos J, Tolosa C, et al. Antimalarial myopathy: an underdiagnosed complication? Prospective longitudinal study of 119 patients. Ann Rheum Dis 2006;65:385-90. [Crossref] [PubMed]
- Stauber WT, Hedge AM, Trout JJ, et al. Inhibition of lysosomal function in red and white skeletal muscles by chloroquine. Exp Neurol 1981;71:295-306. [Crossref] [PubMed]
- Eadie MJ, Ferrier TM. Chloroquine myopathy. J Neurol Neurosurg Psychiatry 1966;29:331-7. [Crossref] [PubMed]
- Mastaglia FL, Papadimitriou JM, Dawkins RL, et al. Vacuolar myopathy associated with chloroquine, lupus erythematosus and thymoma. Report of a case with unusual mitochondrial changes and lipid accumulation in muscle. J Neurol Sci 1977;34:315-28. [Crossref] [PubMed]
- Zirin J, Nieuwenhuis J, Samsonova A, et al. Regulators of autophagosome formation in Drosophila muscles. PLoS Genet 2015;11:e1005006. [Crossref] [PubMed]
- Reichelt ME, Mellor KM, Curl CL, et al. Myocardial glycophagy - a specific glycogen handling response to metabolic stress is accentuated in the female heart. J Mol Cell Cardiol 2013;65:67-75. [Crossref] [PubMed]
- Engel AG. Acid maltase deficiency in adults: studies in four cases of a syndrome which may mimic muscular dystrophy or other myopathies. Brain 1970;93:599-616. [Crossref] [PubMed]
- Griffin JL. Infantile acid maltase deficiency. I. Muscle fiber destruction after lysosomal rupture4. Virchows Arch B Cell Pathol Incl Mol Pathol 1984;45:23-36. [Crossref] [PubMed]
- Raben N, Danon M, Gilbert AL, et al. Enzyme replacement therapy in the mouse model of Pompe disease. Mol Genet Metab 2003;80:159-69. [Crossref] [PubMed]
- Xu S, Galperin M, Melvin G, et al. Impaired Organization and Function of Myofilaments in Single Muscle Fibers from a Mouse Model of Pompe Disease. J Appl Physiol (1985) 2010;108:1383-8. [Crossref] [PubMed]
- Raben N, Shea L, Hill V, et al. Monitoring autophagy in lysosomal storage disorders. Methods Enzymol 2009;453:417-49. [Crossref] [PubMed]
- Raben N, Baum R, Schreiner C, et al. When more is less: excess and deficiency of autophagy coexist in skeletal muscle in Pompe disease. Autophagy 2009;5:111-3. [Crossref] [PubMed]
- Shea L, Raben N. Autophagy in skeletal muscle: implications for Pompe disease. Int J Clin Pharmacol Ther 2009;47 Suppl 1:S42-7. [PubMed]
- Spampanato C, Feeney E, Li L, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 2013;5:691-706. [Crossref] [PubMed]
- Nishino I. Autophagic vacuolar myopathy. Semin Pediatr Neurol 2006;13:90-5. [Crossref] [PubMed]
- Lieberman AP, Puertollano R, Raben N, et al. Autophagy in lysosomal storage disorders. Autophagy 2012;8:719-30. [Crossref] [PubMed]
- Seranova E, Connolly KJ, Zatyka M, et al. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem 2017;61:733-49. [Crossref] [PubMed]
- Hesselink RP, Wagenmakers AJ, Drost MR, et al. Lysosomal dysfunction in muscle with special reference to glycogen storage disease type II. Biochim Biophys Acta 2003;1637:164-70. [Crossref] [PubMed]
- Drost MR, Hesselink RP, Oomens CW, et al. Effects of non-contractile inclusions on mechanical performance of skeletal muscle. J Biomech 2005;38:1035-43. [Crossref] [PubMed]
- Ralston E, Swaim B, Czapiga M, et al. Detection and imaging of non-contractile inclusions and sarcomeric anomalies in skeletal muscle by second harmonic generation combined with two-photon excited fluorescence. J Struct Biol 2008;162:500-8. [Crossref] [PubMed]
- Fukuda T, Ahearn M, Roberts A, et al. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in pompe disease. Mol Ther 2006;14:831-9. [Crossref] [PubMed]
- Fukuda T, Ewan L, Bauer M, et al. Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann Neurol 2006;59:700-8. [Crossref] [PubMed]
- Nascimbeni AC, Fanin M, Tasca E, et al. Impaired autophagy affects acid α-glucosidase processing and enzyme replacement therapy efficacy in late-onset glycogen storage disease type II. Neuropathol Appl Neurobiol 2015;41:672-5. [Crossref] [PubMed]
- Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473-7. [Crossref] [PubMed]
- Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014;7:ra9. [Crossref] [PubMed]
- Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429-33. [Crossref] [PubMed]
- Puertollano R, Ferguson SM, Brugarolas J, et al. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J 2018;37. [Crossref] [PubMed]
- Pastore N, Brady OA, Diab HI, et al. TFEB and TFE3 Cooperate in the Regulation of the Innate Immune Response in Activated Macrophages. Autophagy 2016;12:1240-58. [Crossref] [PubMed]
- Medina DL, Fraldi A, Bouche V, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 2011;21:421-30. [Crossref] [PubMed]
- Settembre C, Ballabio A. Lysosomal Adaptation: How the Lysosome Responds to External Cues. Cold Spring Harb Perspect Biol 2014. [Crossref] [PubMed]
- Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012;31:1095-108. [Crossref] [PubMed]
- Feeney EJ, Spampanato C, Puertollano R, et al. What else is in store for autophagy? Exocytosis of autolysosomes as a mechanism of TFEB-mediated cellular clearance in Pompe disease. Autophagy 2013;9:1117-8. [Crossref] [PubMed]
- Sato Y, Kobayashi H, Higuchi T, et al. TFEB overexpression promotes glycogen clearance of Pompe disease iPSC-derived skeletal muscle. Mol Ther Methods Clin Dev 2016;3:16054. [Crossref] [PubMed]
- Gatto F, Rossi B, Tarallo A, et al. AAV-mediated transcription factor EB (TFEB) gene delivery ameliorates muscle pathology and function in the murine model of Pompe Disease. Sci Rep 2017;7:15089. [Crossref] [PubMed]
- Iwasaki H, Naka A, Iida KT, et al. TFE3 regulates muscle metabolic gene expression, increases glycogen stores, and enhances insulin sensitivity in mice. Am J Physiol Endocrinol Metab 2012;302:E896-902. [Crossref] [PubMed]
- Mansueto G, Armani A, Viscomi C, et al. Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab 2017;25:182-96. [Crossref] [PubMed]
- Lim JA, Sun B, Puertollano R, et al. Therapeutic Benefit of Autophagy Modulation in Pompe Disease. Mol Ther 2018;26:1783-96. [Crossref] [PubMed]
- Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass. Cell Metab 2009;10:507-15. [Crossref] [PubMed]
- Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 2010;6:307-9. [Crossref] [PubMed]
- Wu JJ, Quijano C, Chen E, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425-37. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017;168:960-76. [Crossref] [PubMed]
- Garcia D, Shaw RJ. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol Cell 2017;66:789-800. [Crossref] [PubMed]
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132-41. [Crossref] [PubMed]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014;24:400-6. [Crossref] [PubMed]
- Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496-501. [Crossref] [PubMed]
- Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290-303. [Crossref] [PubMed]
- Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678-83. [Crossref] [PubMed]
- Demetriades C, Plescher M, Teleman AA. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun 2016;7:10662. [Crossref] [PubMed]
- Zhang CS, Jiang B, Li M, et al. The Lysosomal v-ATPase-Ragulator Complex Is a Common Activator for AMPK and mTORC1, Acting as a Switch between Catabolism and Anabolism. Cell Metab 2014;20:526-40. [Crossref] [PubMed]
- Yoon MS. mTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front Physiol 2017;8:788. [Crossref] [PubMed]
- Shemesh A, Wang Y, Yang Y, et al. Suppression of mTORC1 activation in acid-α-glucosidase-deficient cells and mice is ameliorated by leucine supplementation. Am J Physiol Regul Integr Comp Physiol 2014;307:R1251-9. [Crossref] [PubMed]
- Yoshida T, Awaya T, Jonouchi T, et al. A Skeletal Muscle Model of Infantile-onset Pompe Disease with Patient-specific iPS Cells. Sci Rep 2017;7:13473. [Crossref] [PubMed]
- Lim JA, Li L, Shirihai OS, et al. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol Med 2017;9:353-70. [Crossref] [PubMed]
- Xu S, Lun Y, Frascella M, et al. Improved efficacy of a next-generation ERT in murine Pompe disease. JCI Insight 2019. [Crossref] [PubMed]
- Bader CA, Shandala T, Ng YS, et al. Atg9 is required for intraluminal vesicles in amphisomes and autolysosomes. Biol Open 2015;4:1345-55. [Crossref] [PubMed]


