Random forest can accurately predict the development of end-stage renal disease in immunoglobulin a nephropathy patients
Introduction
IgA nephropathy (IgAN), also known as Berger disease, is the most common glomerulonephritis worldwide (1). It is characterized by deposition of IgA in the glomerulus (2) which can lead to end-stage renal disease (ESRD) within 20 years in 40% of patients (3). However, it is currently difficult to predict which IgAN patients will progress to ESRD.
Several studies have explored the relation of demographic (4-8), clinical (4,5,9-12) and pathologic variables (13-18) to predict progression of IgA nephropathy to end stage renal disease. Pathologic evaluation, in particular, has a robust classification scheme, the MESTC score, that has been shown to predict the risk of progression to ESRD. However, MESTC as well as studies of clinical and demographic variables employ standard statistical methods, such as univariate and multivariate Cox regression models, proportional hazards models and cause-specific hazards models which individually only evaluate the relation of a subset of variables to ESRD progression. These limited models potentially ignore important interactions between the variables and their effect on ESRD progression.
Machine learning is a subset of artificial intelligence in the field of computer science that often uses statistical techniques to give computers the ability to “learn” a specific task without being explicitly programmed (19). Many different models fall under the mantle of machine learning, including: logistic regression, random forest, support vector machine (SVM), decision tree, artificial neural network (ANN) and k nearest neighbors (KNN). Such algorithms operate by building a model from an example training set of input observations in order to make data-driven predictions or decisions expressed as outputs, rather than following strictly static program instructions. Decision tree and ANN have previously been used to predict progression to ESRD in IgAN patients (20,21).
In this paper, multiple machine learning models were created (logistic regression, random forest, SVM, decision tree, ANN and KNN) to predict ESRD progression in IgAN patients. The purpose of this study is to successfully identify patients at high risk of progression to ESRD to facilitate early and effective treatment.
Methods
Study cohort
We evaluated data from 1,370 biopsy-proven IgAN patients in West China Hospital, Sichuan University in China between 2009 to 2017. Inclusion criteria included: (I) IgAN diagnosed by a renal biopsy (pathology showed predominant mesangial deposits of IgA by immunofluorescence and mesangial proliferation on light microscopy); (II) patients had complete renal biopsy information and clinical data at the time of kidney biopsy and during follow up; (III) IgAN was the primary disease, not secondary to other diseases; (IV) patients were followed at least 3 months. Patients with the following were excluded from the study: (I) estimated glomerular filtration rate (eGFR) <15 mL/min/1.73 m2 at time of biopsy; (II) had medical history of acute kidney disease.
Dataset collection
After the selection of patients, all demographic, clinical, treatment and pathologic data was extracted, anonymized and stored in an electronic database in Excel format.
Data collected included:
Demographic data: age and gender.
Pathological characteristics: Oxford MESTC score (18). Clinical and laboratory characteristics: systolic blood pressure (SBP), diastolic blood pressure (DBP), hypertension status (HTN), 24-hour urine protein, serum albumin, nephrotic syndrome status (NS), serum creatinine (Scr), serum uric acid (UA), hematuria, eGFR and CKD stage.
Treatment: angiotensin-converting enzyme inhibitors or angiotensin receptor blocker, glucocorticoid and immunosuppressive agents. Endpoint: primary outcome was defined by ESRD. ESRD was defined as the requirement of renal replacement therapy and/or eGFR <15 mL/min/1.73 m2.
Hypertension was defined as arterial blood pressure over 140/90.
Nephrotic syndrome was characterized by proteinuria of >3.5 g per 1.73 m2 body surface area per day.
The eGFR was calculated using the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) formula (22).
CKD was classified using the NKF KDOQI guidelines (National Kidney Foundation Kidney Disease Outcomes Quality Initiative).
Statistics
The demographic, clinical, treatment and pathologic characteristics at the time of biopsy were retrospectively compared between patients who progressed to ESRD (ESRD group) and those who did not progress to ESRD (non-ESRD group). Continuous data were reported as mean and SD for normal distributions and as median and interquartile range for non-normal distributions. The categorical data were reported as number and percentage. The t-test was used for continuous variables, and Pearson Chi-Square test for categorical variables. All tests of significance were 2-tailed and the P<0.05 were considered statistically significant. The IBM SPSS Statistics software (Version 20.0. IBM Corporation, NY, USA) was used for analyses.
Mathematical model
Seventy-five percent of all patients were randomly selected to be analyzed to create a mathematical model to predict the status of ESRD, and 25% were used to test the mathematical model. Univariate logistic analysis was used to identify all parameters affecting the status of ESRD and multivariate logistic analysis was performed to select independent prediction factors. A mathematical model for predicting the status of ESRD was created based on the results of the multivariate logistic analysis. The efficacy of MM was defined as accuracy, sensitivity, specificity and AUC. P<0.05 were considered statistically significant. The IBM SPSS Statistics software (Version 20.0. IBM Corporation, NY, USA) was used for analyses.
Supervised learning classifiers
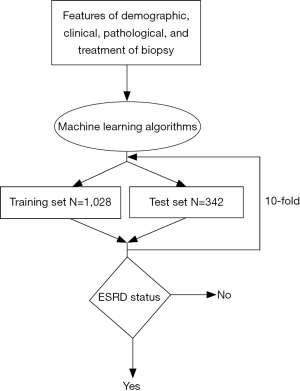
Random forest was applied along with other five classifiers (logistic regression, support vector machine, decision tree, ANN and KNN) to predict the status of ESRD. All factors were enrolled in the prediction models. The status of ESRD was classified as “negative” or “positive”. Thus, the prognosis of ESRD status is a binary classification task. The efficacy of the models was defined as four key metrics: overall accuracy, sensitivity, specificity, and AUC (area under curve). Each metric was tested under 10-fold cross validation that randomly selected 75% of the dataset as the training set (n=1,028) and the rest as the test set (n=342) (Figure 1).
Results
Patients characteristics
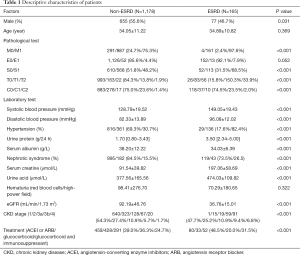
Twenty-seven patients were excluded because of AKI and/or eGFR <15 mL/min/1.73 m2 at time of biopsy. A total of 1,343 patients were eventually included ranging from 14 to 64 years old. The average age of this cohort was 34.15±11.17 years old and mean follow up was 45.5±23.98 months. The demographic description, laboratory test characteristics, kidney biopsy results and treatment at biopsy of the cohort are shown in Table 1. 165 patients were diagnosed of ESRD at an average following time of 34.8 months. Males comprised 55.6% and 46.7% in the non-ESRD and ESRD groups, respectively. Patients’ ages between the two groups were not significantly different. All pathological scores, except for the E score from the biopsy were highly related to progression to ESRD. All laboratory tests demonstrated significance except for the presence of hematuria.
Full table
Mathematical model
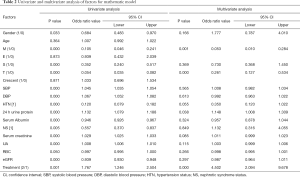
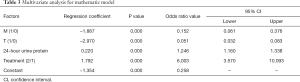
As shown in Table 2, the following factors were related to univariate analyses: gender, M score, S score, T score, SBP, DBP, HTN, 24 h urine protein, serum albumin, NS, Scr, UA, hematuria, and treatment. Of these, M score, T score, 24 h urine protein and treatment were independent factors for ESRD status after multivariate analysis. The mathematical model was devised based on the multivariate analysis of independent factors (Table 3). The following formula was employed: P = eX/(1+eX) where X= –1.354–(1.887*M) – (2.970*T) + (0.22*24 h urine protein) + (1.792*treatment), e is the base of the natural logarithm. For M and T score, 0 and 1 were defined as absent and present. For treatment, 1 and 2 were defined as no glucocorticoid use and glucocorticoid use. A P value of 0.645 was selected as a cut-off point and P value >0.645 should be considered positive ESRD status and P<0.645 should be considered negative.
Full table
Full table
The mathematical model was tested by test group, and performed the accuracy of 43.62%, sensitivity of 10.77%, and specificity of 51.47% in Table 2. The area under ROC curve of this model was 34.4%, which is not shown in the present study.
ESRD status predictions
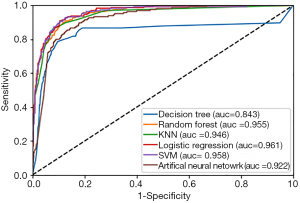
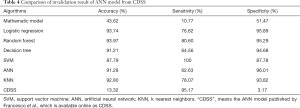
Six supervised classification models were generated to predict progression to ESRD. Table 4 compared the detail performance of the cross-validation results of each algorithm. As can be seen from Table 4, random forest obtained highest accuracy of 93.97%, with sensitivity of 80.60% and specificity of 95.29%. SVM achieved 100% sensitivity and ANN has 96.01% specificity, which are outperformed other algorithms.
Full table
Figure 2 demonstrates the receiver operating characteristic (ROC) curves of the six prediction models. Logistic regression [area under the curve (AUC) =96.1%] outperformed the other models [random forest (AUC =95.5%), SVM (AUC =95.8%), decision tree (AUC =84.3%), ANN (AUC =92.2%) and KNN (AUC =94.6%)].
Discussion
Although up to 40% IgAN patients will progress to ESRD, it is difficult to predict which patients will and which won’t progress. A useful and practical method, which could accurately and quickly estimate a patient`s long-term renal outcome would be useful for nephrologists to improve clinical decision making.
Most baseline factors of patients at onset between negative and positive ESRD groups are significantly different so it is possible to use baseline factors at onset to predict long-term ESRD status of IgAN patients. Conventional mathematic prediction model and machine learning methods were both utilized in our study. We demonstrated that machine learning can achieve much higher predictive accuracy than a typical mathematical model in our cohort. Only 4 factors were enrolled in the previous mathematical model, which is much less than 19 factors used with machine learning models. Comparing 6 machine learning models, random forest outperformed the other 5 algorithms. Randon forest is a classifier that contains multiple decision trees and the categories it outputs are determined by the number of classes of individual tree outputs. When determining the category, random forest can assess the importance of all variables. Random forest can handle a large number of input variables and is able to estimate lost data to maintain the accuracy.
A lot of factors were used in our cohort, especially the pathological characteristics. MESTC score were last updated in 2017, and were the diagnostic factors of IgAN. Despite their important diagnostic and clinical significance, few studies have discussed whether MESTC score can predict the likelihood of progression to ESRD in IgAN patients. Tanaka (16), Barbour (15) and Xie (17) all showed that MEST score could be predictive. While, Pesce (21) used another pathological system to convey overall severity of the lesions found in each tissue compartment rather than 4 MEST lesions. Zhang (23) recently demonstrated crescents, scored as “0”, “1”, “2”, were significant in predicting ESRD in Chinese IgAN patients. Although treatment at baseline does not reflect the patient`s severity, effects of treatment on the prognosis of IgA nephrology could be evaluated when adding to the prediction models. Our previous study showed combined immunosuppressive treatment could improve short renal outcome in advanced IgAN compared to steroids alone (24). Therefore, our model took all 19 available features (gender, age, SBP, DBP, HTN, 24-hour proteinuria, serum albumin, nephrotic syndrome status, serum creatinine, urine acid, hematuria, eGFR and CKD stage, MEST score, crescent score and treatment at baseline) as the input, achieving highest accuracy with 93.97% when using random forest.
Some papers also showed the application of machine learning algorithms in IgA nephrology prediction. The strength of our study compared with others is that we use all possible parameters as input to avoid ignoring non-statistically-significant parameters. Pesce et al. (21) created an ANN model available online, called as CDSS (clinical decision support system), taking only 6 features (gender, age, histological grading, serum creatinine, proteinuria and HTN) into account with an accuracy of 91.8%, which was lower than our random forest system. The CDSS system applied to our cohort demonstrated an accuracy of 13.32%, specificity of 3.17%, which are low, but with high sensitivity of 95.17% (Table 4). That may be because many negative ESRD patients in our database were treated as positive in CDSS. Perhaps an additional factor in the low performance of CDSS in our cohort is the use of histological grading other than the MEST score which is the worldwide accepted classification system for IgA nephropathy pathological grading. A decision tree was created by Goto et al. (20) with a ROC of 83.0%, which is lower than our random forest model (95.5%). Random forest can handle large numbers of predictors and incorporate different types of features which is common in patients’ characteristics.
Some limitations of this study must be acknowledged. Firstly, only Chinese patients were included in the model, prediction for other populations was not evaluated. Secondly, our cohort was not large enough and had lost and unbalanced data. Lastly, out study is a retrospective study which could not show the effect of the model on directing treatment. Nonetheless, the current prediction model is an effective and simple method to predict progression of IgA nephropathy patients to ESRD.
In conclusion, in IgAN patients without severe impairment of renal function, the status of progression to ESRD can be easily predicted with clinical and pathological information by random forest prediction model with high accuracy. In future work, large and multicenter data are needed for establishing prediction models as prospective study.
Acknowledgments
Funding: The work of Ying Wang was partially supported by the NSF grants DMS-1720489 and DMS-1752709.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The research was in compliance of the Declaration of Helsinki and was approved by the ethical committee of West China Hospital of Sichuan University.
References
- D'Amico G. The commonest glomerulonephritis in the world: IgA nephropathy. Q J Med 1987;64:709-27. [PubMed]
- Berger J, Hinglais N. Intercapillary deposits of IgA-IgG. J Urol Nephrol (Paris) 1968;74:694-5. [PubMed]
- Manno C, Strippoli GF, D'Altri C, et al. A novel simpler histological classification for renal survival in IgA nephropathy: a retrospective study. Am J Kidney Dis 2007;49:763-75. [Crossref] [PubMed]
- Radford MG Jr, Donadio JV Jr, Bergstralh EJ, et al. Predicting renal outcome in IgA nephropathy. J Am Soc Nephrol 1997;8:199-207. [PubMed]
- Goto M, Wakai K, Kawamura T, et al. A scoring system to predict renal outcome in IgA nephropathy: a nationwide 10-year prospective cohort study. Nephrol Dial Transplant 2009;24:3068-74. [Crossref] [PubMed]
- Eriksen BO, Ingebretsen OC. The progression of chronic kidney disease: a 10-year population-based study of the effects of gender and age. Kidney Int 2006;69:375-82. [Crossref] [PubMed]
- Ouyang Y, Xie J, Yang M, et al. Underweight Is an Independent Risk Factor for Renal Function Deterioration in Patients with IgA Nephropathy. PLoS One 2016;11:e0162044. [Crossref] [PubMed]
- Ruggajo P, Svarstad E, Leh S, et al. Low Birth Weight and Risk of Progression to End Stage Renal Disease in IgA Nephropathy--A Retrospective Registry-Based Cohort Study. PLoS One 2016;11:e0153819. [Crossref] [PubMed]
- Xie J, Kiryluk K, Wang W, et al. Predicting progression of IgA nephropathy: new clinical progression risk score. PLoS One 2012;7:e38904. [Crossref] [PubMed]
- Berthoux F, Mohey H, Laurent B, et al. Predicting the risk for dialysis or death in IgA nephropathy. J Am Soc Nephrol 2011;22:752-61. [Crossref] [PubMed]
- Wakai K, Kawamura T, Endoh M, et al. A scoring system to predict renal outcome in IgA nephropathy: from a nationwide prospective study. Nephrol Dial Transplant 2006;21:2800-8. [Crossref] [PubMed]
- Donadio JV, Bergstralh EJ, Grande JP, et al. Proteinuria patterns and their association with subsequent end-stage renal disease in IgA nephropathy. Nephrol Dial Transplant 2002;17:1197-203. [Crossref] [PubMed]
- Roberts ISD, Cook HT, Troyanov S, et al. The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int 2009;76:546-56. [Crossref] [PubMed]
- Working Group of the International IgA Nephropathy Network, Lu SE, Lin Y, et al. The Oxford classification of IgA nephropathy: rationale, clinicopathological correlations, and classification. Kidney Int 2009;76:534-45. [Crossref] [PubMed]
- Barbour SJ, Espino-Hernandez G, Reich HN, et al. The MEST score provides earlier risk prediction in lgA nephropathy. Kidney Int 2016;89:167-75. [Crossref] [PubMed]
- Tanaka S, Ninomiya T, Katafuchi R, et al. Development and validation of a prediction rule using the Oxford classification in IgA nephropathy. Clin J Am Soc Nephrol 2013;8:2082-90. [Crossref] [PubMed]
- Xie J, Lv J, Wang W, et al. Kidney Failure Risk Prediction Equations in IgA Nephropathy: A Multicenter Risk Assessment Study in Chinese Patients. Am J Kidney Dis 2018;72:371-80. [Crossref] [PubMed]
- Haas M, Verhave JC, Liu ZH, et al. A Multicenter Study of the Predictive Value of Crescents in IgA Nephropathy. J Am Soc Nephrol 2017;28:691-701. [Crossref] [PubMed]
- Langley P. The changing science of machine learning. Machine Learning 2011;82:275-9. [Crossref]
- Goto M, Kawamura T, Wakai K, et al. Risk stratification for progression of IgA nephropathy using a decision tree induction algorithm. Nephrol Dial Transplant 2009;24:1242-7. [Crossref] [PubMed]
- Pesce F, Diciolla M, Binetti G, et al. Clinical decision support system for end-stage kidney disease risk estimation in IgA nephropathy patients. Nephrol Dial Transplant 2016;31:80-6. [Crossref] [PubMed]
- Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604-12. [Crossref] [PubMed]
- Zhang X, Shi S, Ouyang Y, et al. A validation study of crescents in predicting ESRD in patients with IgA nephropathy. J Transl Med 2018;16:115. [Crossref] [PubMed]
- Tan L, Tang Y, Peng W, et al. Combined Immunosuppressive Treatment May Improve Short-Term Renal Outcomes in Chinese Patients with Advanced IgA Nephropathy. Kidney Blood Press Res 2018;43:1333-43. [Crossref] [PubMed]