
Methylation of TP53BP2 and Apaf-1 genes in embryonic lung cells and their impact on gene expression
Introduction
Lungs are internal organs related to human respiration. In recent years, due to air pollution (e.g., smog) and other human factors (smoking, occupational hazards, etc.), lung-related diseases such as lung cancer (1), tuberculosis (2) continue to emerge; Meanwhile, internal factors such as genetic, epigenetic or familial factors are also important causes of lung disease. Epigenetics plays an irreplaceable role during embryonic development in maintaining the normal life activities of mammals (3), in which the most important pattern is the rearrangement of DNA methylation in primordial germ cells and embryonic cells (4). Therefore, the study of methylation during embryonic lung development will gain a better understanding of the pathogenesis of lung disease. In studies of DNA methylation, bioinformatics can not only analyze large amounts of data generated by high-throughput detection technologies, but also predict methylation-related information in specific genomic regions, which can supply valuable reference and guidance for experimental research. CpG islands are useful markers for genes in organisms containing 5-methylcytosine in their genomes. In addition, CpG islands located in the promoter regions of genes can play important roles in gene silencing. We pursued study on the CpG islands of two important genes, TP53BP2 (tumor protein p53 binding protein 2) and Apaf-1 (apoptotic protease activating factor-1) of human fetal lung fibroblasts (HFL-I). TP53BP2 gene encodes p53 binding protein 2 (53BP2) which promotes apoptosis through the coupling of important regulators p53/p73 pathway as well as Rb/E2F pathway. The Apaf-1 may play an important role in DNA damage-induced apoptosis, which is a transcriptional target of E2F and p53. In this study, based on bioinformatics predictions combined with the analysis of bisulfite sequencing PCR (BSP) (5) as well as gene transcription regulation, the distribution and extent of methylation of embryonic lung cells were studied and DNA methylation and its effect on gene expression regulation were explored.
Methods
Bioinformatics prediction of TP53BP2 and Apaf-1 genes (dry experiment)
CpG island in promoter region
The original gene sequences of TP53BP2 and Apaf-1 were analyzed. Sequences of 2,000 bp (from upstream of the transcription initiation site to downstream of the transcription initiation site) in promoter areas were download from NCBI (https://www.ncbi.nlm.nih.gov/) and the bioinformatics predictions of promoter-related CpG islands were analyzed by CpG Island Searcher (6) and Methprimer (7) program on line (Table 1). The parameter settings are as follows: (I) CpG Island Searcher program, the lower limit values: GC% =55%, ObsCpG/ExpCpG =0.65, length =500 bp; (II) Methprimer program: island size >100, GC percent >50.0, Obs/Exp >0.60.
Full table
TF binding sites (TFBS) in CpG island of promoter region
To investigate the possible relationship between the methylation of CpG islands in promoter regions of TP53BP2 and Apaf-1 and the binding of transcription factors (TFs), the gene sequences in CpG island above were predicted by Match program (8) (the thresholds for degrees and core similarities were 0.9 and 0.95, respectively), TFSEARCH program based on the TRANSFAC databases (9), and Promoter Scan program (10) to predicting the TFBS (Table 1).
Methylation detection of TP53BP2 and Apaf-1 genes in CpG island (wet experiment)
Culture and drug treatment of cells
HFL-I cells were purchased from the China Center for Type Culture Collection (Shanghai, China) and cultured according to the manufacturer’s instructions. For 5-aza-2'-deoxycytidine (5-aza-CdR, Sigma) treatments, cells were diluted to 3.0×105/mL and allowed to grow overnight; then freshly prepared 5-aza-CdR was added to the cells at a final concentration of 3 µM and the cells were cultured for up to 72 h. Fresh medium containing 5-aza-CdR was added every 24 h. Cells were then harvested for genomic DNA (gDNA) and RNA isolation.
Preparation of gDNA, total RNA
gDNA was isolated with AxyPrepTM Blood Genomic DNA Midiprep Kit (Axygen Scientific), according to the manufacturer’s protocols. Total RNA from cells was extracted using TRIzol reagent (TaKaRa) as the manufacturer's instructions. The quality and quantity of the extracted gDNA and total RNA were evaluated by agarose gel electrophoresis and NanoDrop® ND-1000 Spectrophotometer (Thermo Fisher), respectively.
Detection of DNA methylation with BSP
The gDNA samples of HFL-I were subjected to bisulfite modification by using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s recommendations. The promoter regions of TP53BP2 and Apaf-1 were amplified from the bisulfite-treated gDNA with the primers of TP53BP2-Fm/Rm and Apaf-1-Fm/Rm listed in Table 2. The PCR reaction consisted of 15 µL 2× PCR Mixture, 1–5 µL gDNA, 0.5 µL 10 µM forward primer (Fm), 0.5 µL 10 µM reverse primers (Rm), and 9–13 µL water. PCR products were detected by 1.8% (w/v) agarose gel electrophoresis. Bisulfite sequencing were performed with at least 10 individual clones by using ABI3730 automatic DNA sequencer (Applied Biosystems). The promoter-associated CpG islands were searched online with CpG Island Searcher using the NCBI original sequences. The results of BSP were analyzed online with BISMA (Bisulfite Sequencing DNA Methylation Analysis) software in FASTA format (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/index.php).

Full table
Detection of gene expression with quantitative PCR
Complementary DNA (cDNA) was synthesized from 1 µg of total RNA as a template using a Reverse Transcriptase Kit (TaKaRa), following manufacturer’s instructions. Quantitative PCR was performed with Fast SYBR Master Mix (Applied Biosystems, UK) using primers listed in Table 2 on a StepOneTM Plus instrument (Applied Biosystems). The PCR reaction consisted of 10 µL 2× mix of Fast SYBR Master Mix, 1 µL cDNA, 0.4 µL 10 µM forward primer (Table 2), 0.4 µL 10 µM reverse primers (Table 2), and 8.2 µL water. The PCR reaction was: 95 °C 10 min; 40 circles of 95 °C 15s, 58 °C (TP53BP2 and GAPDH) or 60 °C (Apaf-1) 1 min; and 72 °C 5 min. A melting curve stage was performed after the completion of PCR. The expression of the target genes above was normalized against a housekeeping gene (GAPDH).
Statistical analysis
Using statistical analysis software for data processing. The t-test method was used to compare the difference among the groups. P<0.05 was considered to be statistically significant difference.
Results
Fluorescence micrographs of HFL-I are shown in Figure 1. From the micrographs, the morphology of HFL-I cells were normal. After staining with DAPI dyestuff, the cell nuclei showed blue fluorescence (Figure 1C).
Bioinformatics predictions of TP53BP2 and Apaf-1 genes
Prediction of CpG island in promoter region
As the gene promoter region is usually located from 2,000 bp upstream to 200 bp downstream of transcription start site (TSS), CpG island predictions of short sequences from 1,000 bp upstream to 1,000 bp downstream of TSS (TP53BP2 and Apaf-1 genes) were performed using MethPrimer software on line. The results are shown in Figure 2, the TSS are located at 1,000 bp. The distribution of CG dinucleotide in these regions near TSS are dense. These regions are usually considered as important regulatory domains, i.e., the proximal promoter region. Therefore, it was inferred that TF binding and gene expression regulation in these regions may be affected by methylation. In addition, TP53BP2 gene showed an independent CpG island, while the CpG island of the Apaf-1 gene showed a discontinuous state (CpG island in blue of Figure 2A,B). In conclusion, the predictions of CpG island indicate that TP53BP2 and Apaf-1 are typical genes that containing the CpG islands.
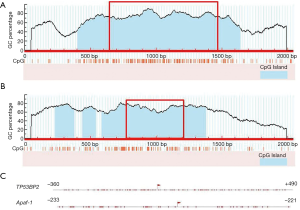
Based on the results above, the sequences from proximal promoter regions of TP53BP2 and Apaf-1 genes (850 and 455 bp, respectively) shown in the red box in Figure 2 were cut off, and these areas of CpG island were predicted furtherly using another software online: CpG Island Searcher (Figure 2C). The results showed that the two predictions are consistent. So genomic sequences of 850 and 455 bp above of TP53BP2 and Apaf-1 genes respectively were selected for further prediction, detection and analysis.
Prediction of TFBS in CpG island
The predicted CpG islands in promoter regions of TP53BP2 and Apaf-1 genes are mainly distributed in proximal promoter regions, which are also important binding regions for TF. TFBS prediction and sequences analysis of 850 and 455 bp selected above were performed using online programs of Match program, TFSEARCH, and Promoter Scan. As shown in Figure 3, the CpG island regions in proximal promoter regions of both genes contain multiple TFBS, such as E2F, GATA-1, c-Ets-1, AREB6, CREB, USF, Sp1, Ik-1, Nkx2-5, AML-1a, Oct-1, NF-κB, AP-1, SRY, etc.; Some TFs contain multiple binding sites, such as E2F, GATA-1, c-Ets-1, AREB6, CREB, USF, Sp1 and Ik-1. Among them, the E2F-TFBS appeared 5 times in the TP53BP2 gene and 4 times in the Apaf-1 gene; the CREB-TFBS appeared 3 times in the TP53BP2 gene, and c-Ets-1-TFBS appeared 4 times in two genes.

Then the putative methylation CpG sites were determined by Methylator program (11) on line. The results showed that parts of the CpG dinucleotides were methylated. For example, the TP53BP2 gene had a total of 121 CpGs, and 61 methylated CpG dinucleotides were found. In addition, these CpG dinucleotides prone to methylation also appeared in some important TFBS such as E2F, Sp1, AREB6, CREB, AP-1, AML-1α, Sp1, ATF, GATA-1, c-Ets-1, USF, Ik-1, etc.; some TFBS also contained multiple methylated CpG dinucleotides, such as E2F, AP-1, USF, ATF, GATA-1, and AhR/Arnt.
Methylation detection of CpG island in promoter region of TP53BP2 and Apaf-1 genes
To investigate the accuracy of bioinformatics predictions and the relationship between methylated CpG at TFBS and target gene expression of TF, the 850 and 455 bp of the proximal promoter regions of TP53BP2 and Apaf-1 were conducted by BSP to detect the methylation levels of both genes, respectively.
BSP
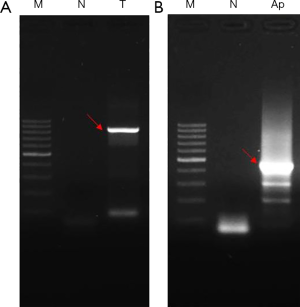
Sequencing after bisulfite treatment is the gold standard method for the detection of methylation status of specific gene sites (12). The rationale is that the unmethylated cytosine (C) becomes uracil (U) after bisulphite treatment, while the methylated cytosine (mC) remains unchanged. After PCR amplification, the uracil at the unmethylated site is amplified as thymine (T), while the cytosine at the methylation site remains in cytosine form (mCG). The difference can be detected by sequencing. Here, the genomic DNA of HLF-I cells was subjected to bisulfite treatment, and PCR amplifications were performed aiming at the CpG islands of promoter region of TP53BP2 and Apaf-1 genes. The results are shown in Figure 4, two target DNA fragments are successfully amplified (indicated by red arrows), indicating that the successful bisulfite treatments were performed.
After obtaining the two target DNA fragments, pMD19T (TaKaRa) was used as vector plasmid followed by transformation, resuscitation, plating and PCR amplification. Ten positive clones were selected for inoculation and cultured overnight at 37 °C. Plasmid extraction was performed using a plasmid extraction kit. Two plasmids were selected for double enzyme digestion to determine the insertion of the fragments. The plasmid extraction results are shown in Figure 5 by agarose gel electrophoresis and DNA target fragments were sequenced using the ABI3730 automated DNA sequencer.
BISMA results
The BSP results of 10 clones were visualized in FASTA format using BISMA software (Figure 6). As can be seen from the figure, TP53BP2 gene has a total of 120 CpG sites, of which 10, 116–118 CpG sites are detected methylation in 3 out of 10 clones, the methylation percentage (CpG%) is 30%; CpG sites 65, 67, 69, 71–73, 79, 84 and 101 are detected methylation in 2 out of 10 clones, the percentage of methylation is 20%; The CpG sites of 3, 6, 7–9, 12, 14, 18, 19, 22, 33, 36, 45, 48, 52, 59, 60, 77–78, 86, 87, 91, 94, 98, 100 and 112 are detected methylation in 1 out of 10 clones, the percentage is 10%.
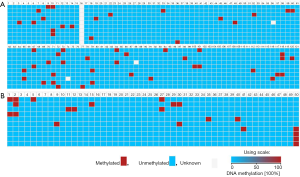
The Apaf-1 gene has a total of 50 CpG sites. The percentage of CpG methylation at each site are as follows: the CpG site of 50 is 40%; the CpG site of 2 is 30%; 1, 27, 30 sites are 20%; 5, 9, 11, 12, 15, 29, 35, 40, and 41 are 10%. These methylated CpGs also appear at the binding sites of partial TFs (TF-DBS). These TF-DBSs include ARFB6, AP-1, E2F, AML-1α(+)/PEBP2, USF, CREB, c-Ets-1, Hand1/E47, SP1, GATA-1, Ik-1, and Elk-1 of TP53BP2 gene and NF-Y, AhR/Arnt, c-Ets-1, Hand1/E47, etc. of Apaf-1 gene. In addition, comparing the sequencing results with bioinformatics predictions, it is found that methylated CpGs partially overlap the methylation-prone CpGs predicted by bioinformatics methods and occurs in some important TF-DBSs, such as TP53BP2 gene: the TFs and their CpG site numbers are as follows: AREB6 [6], AP-1 [7], E2F [48, 71, 72], AML-1α (+)/PEBP2 [19], USF [52], CREB [65], c-Ets-1 [91], SP1 [67], GATA-1 [98], Ik-1 [112], NF-κB (+)/c-Rel [118] and Elk-1 [120].
Detection of gene transcription with quantitative PCR
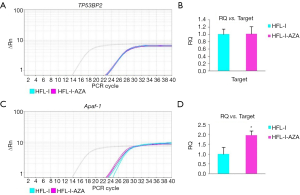
To investigate the methylation of proximal promoter region of CpG island and its effect on the regulation of gene expression, the transcription of TP53BP2, Apaf-1 and GAPDH in HFL-I cells before and after treatment with the methylation inhibitor 5-aza-CdR were detected with qPCR. The results are shown in Figure 7. The transcription of Apaf-1 was significantly increased compared with TP53BP2 after drug treatment. The level of gene expression of Apaf-1 was higher than that of before drug treatment (P<0.05); There was no significant changes of TP53BP2 gene before and after treatment (P>0.05).
Discussion
Studies have shown that epigenetics, especially DNA methylation, is closely related to the establishment of gene imprinting, regulation of gene expression and morphogenesis of embryos during embryonic development. The DNA methylation mechanism and its role in embryonic development have been extensively studied by different biological models such as human (13), mouse (14), zebrafish (15).
In this study, we investigated the DNA methylation and its regulation of gene expression in human embryonic lung cells (HFL-I). By bioinformatics predictions of CpG islands in promoter regions of TP53BP2 and Apaf-1, it was found that the CpG dinucleotide of the two genes were closely spaced and located close to gene TSS; These two regions were also terminal promoter regions, it was speculated that methylation modification in this region may affect TF binding and regulation of related gene expression. By predicting the TFBS in promoter regions of the two genes, we found dense TFBS in these regions, such as AREB, c-Ets-1, E2F, AP-1, Elk-1, GATA-1, Hand1/E47, AML1 and Sp1, etc., suggest that TP53BP2 and Apaf-1 may be subject to complex regulation of multiple TFs. Among them, the E2F-TFBS shows multiple times in the proximal promoter region. The BSP results of key regions containing the E2F-TFBS also reveal that the methylated CpG partially overlap with the predicted methylation-prone CpGs of the two genes and appear at some important TFBS.
Previous studies have shown that E2F is an important TF family. The DNA binding site of this family member contains highly conserved CpG. The E2F TF is also a typical TF affected by CpG methylation. TP53BP2 gene studied here can promote apoptosis through the coupling of important regulators p53/p73 pathway as well as Rb/E2F pathway, which is an important E2F target gene (16). The Apaf-1 gene may play an important role in DNA damage-induced apoptosis (17), which is also a transcriptional target of E2F and p53. Both of genes above play an important role in apoptosis, they also show different degree of methylation in early stages of embryonic development.
We found that after methylation inhibitor intervention, the transcription of Apaf-1 was significantly increased compared with TP53BP2, suggesting that partial methylation in proximal promoter of Apaf-1 have a certain effect on transcription Inhibition, and hypermethylation inhibited the transcription of this gene. The demethylation of the promoter region could increase the transcription of this gene in HFL-I cells. However, compare to primary cell, the methylation of HFL-I cell line may change during the establishment of a strain, which may be one of the reasons that upregulation is not obvious of TP53BP2 transcript.
In addition to E2F, other TFs such as SP1, CREB affected by CpG methylation are also appear in the proximal promoter region of these two genes. Studies support that co-regulation of multiple TFs may be an important gene regulation mechanism in eukaryotes (18). It is also a mechanism to control gene expression in different tissues and cells with different spatio-temporal characteristics. This mechanism is especially significant for cells and tissues during embryonic development. However, the methylation in promoter regions of TP53BP2 and Apaf-1 as well as the distribution of TFBS play an important role in co-regulation of multiple TFs and cell reprogramming in embryonic development. Bioinformatic prediction (dry experiment) combined with BSP and qPCR (wet experiment) can help to learn more about the epigenetic regulation mechanism of embryonic lung and its impact on the embryonic development.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant No. 81702103), Jiangsu Provincial Natural Science Foundation (grant No. BK20170252), Projects for Jiangsu Provincial Young Medical Talents (grant No. QNRC2016780), General Program of the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant No. 16KJD320005), and Xuzhou Science and Technology Planning Project (grant No. KC16SY157).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Wang GZ, Cheng X, Li XC, et al. Tobacco smoke induces production of chemokine CCL20 to promote lung cancer. Cancer Lett 2015;363:60-70. [Crossref] [PubMed]
- Sun J, Wang J, Wei Y, et al. The Haze Nightmare Following the Economic Boom in China: Dilemma and Tradeoffs. Int J Environ Res Public Health 2016;13:402. [Crossref] [PubMed]
- Jiang L, Qin L, Li MJ. Research progress in human embryonic development, spermatogenesis and DNA methylation. Medical Recapitulate 2013:197-9.
- Walter M, Teissandier A, Pérez-Palacios R, et al. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife 2016;5. [Crossref] [PubMed]
- Miyata K, Naito M, Miyata T, et al. Bisulfite Sequencing for DNA Methylation Analysis of Primary Muscle Stem Cells. Methods Mol Biol 2017;1668:3-13. [Crossref] [PubMed]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol 2003;3:235-40. [PubMed]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427-31. [Crossref] [PubMed]
- Kel AE, Gössling E, Reuter I, et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 2003;31:3576-9. [Crossref] [PubMed]
- Heinemeyer T, Wingender E, Reuter I, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 1998;26:362-7. [Crossref] [PubMed]
- Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol 1995;249:923-32. [Crossref] [PubMed]
- Bhasin M, Zhang H, Reinherz EL, et al. Prediction of methylated CpGs in DNA sequences using a support vector machine. FEBS Lett 2005;579:4302-8. [Crossref] [PubMed]
- Shafi A, Mitrea C, Nguyen T, et al. A survey of the approaches for identifying differential methylation using bisulfite sequencing data. Brief Bioinform 2018;19:737-53. [Crossref] [PubMed]
- Li J, Han W, Shen X, et al. DNA methylation signature of long noncoding RNA genes during human pre-implantation embryonic development. Oncotarget 2017;8:56829-38. [PubMed]
- Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010;463:1101-5. [Crossref] [PubMed]
- Tittle RK, Sze R, Ng A, et al. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol 2011;350:50-63. [Crossref] [PubMed]
- Chen D, Padiernos E, Ding F, et al. Apoptosis-stimulating protein of p53-2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ 2005;12:358-68. [Crossref] [PubMed]
- Norbury CJ, Zhivotovsky B. DNA damage-induced apoptosis. Oncogene 2004;23:2797-808. [Crossref] [PubMed]
- Van Roey K, Davey NE. Motif co-regulation and co-operativity are common mechanisms in transcriptional, post-transcriptional and post-translational regulation. Cell Commun Signal 2015;13:45. [Crossref] [PubMed]


