
Host-pathogen interactions in typhoid fever: the model is the message
Salmonella enterica subspecies enterica is classified into serovars based on serological identification of lipopolysaccharide (O) and flagellar (H) antigens (1). Depending on the serovar and the host, Salmonella infection of humans has different outcomes that can be classified into two general types: (I) gastroenteritis, a self-limited infection of the terminal ileum and colon leading to diarrhea and inflammation, often caused by the broad-range serovars Typhimurium and Enteritidis; (II) typhoid fever, a systemic infection caused by the human-adapted serovars Typhi and Paratyphi. Both types of infection usually occur upon ingestion of contaminated food or water. Gastroenteritis, popularly known as salmonellosis, is confined to the intestine and rarely presents complications in healthy adults. In typhoid fever, Salmonella cells disseminate through the lymphatic system and within phagocytes, leading to colonization of internal organs such as the liver, the spleen, the bone marrow, and the gall bladder (2). Infection by S. Typhi may remain asymptomatic for 1–2 weeks, and its symptoms vary from one infected individual to another, depending on age, immune proficiency, and other factors. Common symptoms include high fever, constipation, headache, abdominal tenderness, coughing, and vomiting. Serious complications of typhoid fever are intestinal bleeding, sometimes with perforation, and typhoidal encephalopathy (2). Outbreaks of typhoid fever occur mostly in developing countries, especially in areas with a high population density, and the number of cases per year may be as high as 10–20 million worldwide, resulting in 10,000–20,000 deaths. A fraction of typhoid fever survivors (2–5%) become chronic carriers of S. Typhi (3). The main niche for chronic carriage is the gall bladder, especially in patients with gallstones. A potential link between chronic carriage and hepatobiliary cancer has been considered (3,4). As a consequence of gall bladder contraction during food passage, bacterial shedding into the intestine of chronic carriers occurs, with concomitant release of S. Typhi into the environment.
Over several decades, studies with animal models and cell cultures have unraveled mechanisms employed by Salmonella serovars to cause disease in humans. A specially successful animal model has been infection of immunodeficient mice (BALB/c and other breeds) with S. Typhimurium, which causes an illness somewhat similar to human typhoid fever (5). An obvious advantage of these popular models is the avoidance of the cumbersome biosafety constraints involved in work with typhoidal serovars. However, discrepancies between the S. Typhimurium model and the S. Typhi infection have accumulated in the last decade, and have been comprehensively reviewed recently (6). Differences in genome content, gene regulation and activity of virulence effectors illustrate the limitations of S. Typhimurium-based models of typhoid (6). As a consequence, crucial aspects of S. Typhi infection remain to be understood, thus limiting the efficacy of diagnostic and therapeutic procedures as well as the development of reliable vaccines.
In a recent study, the early stages of S. Typhi infection have been investigated in intestinal biopsies and in organoids derived from human intestinal epithelial tissue (7). Infected biopsies were used to monitor transcriptional changes in the host and the pathogen, to determine cytokine profiling, and to perform electron microscopy observation. In turn, specific mechanisms were investigated using an organoid model. Unlike traditional epithelial cell cultures growing as monolayers on a plate, organoids mirror the in vivo organization of the intestinal epithelium as they develop a three-dimensional architecture (8). Organoids contain the different cell types that constitute the intestinal epithelium (9), which is a major line of defense against microbial pathogens due to the abundance of immune system components (10). Such components are present in organoids and not in epithelial cell cultures.
Active immune evasion by Salmonella Typhi
Transcriptomic analysis in biopsy cells infected with S. Typhi revealed downregulation of human loci involved in B-cell receptor signaling and in coordination between innate and adaptive immune responses. In contrast, few human genes were found to be upregulated during infection. Interestingly, comparison of expression patterns in biopsies infected with either S. Typhi or S. Typhimurium showed stronger downregulation of host genes upon S. Typhi infection. Monitoring of MAPK and NF-κB signaling cascades triggered by bacterial invasion also provided evidence that S. Typhimurium elicits a stronger immune response. Cytokine release was detected in supernatants from biopsies infected by S. Typhi; however, cytokine genes were not found to be upregulated during infection, suggesting that exposure to S. Typhi reduces transcription of cytokine genes. The observation that S. Typhimurium elicits a stronger immune response may help to understand the capacity of S. Typhi to invade deep tissues, and confirms previous evidence that S. Typhi disarms early innate and adaptive immune responses (Figure 1). For instance, the regulatory protein TviA, encoded within the S. Typhi-specific pathogenicity island SPI-7, has been shown to attenuate the innate immune response by downregulating bacterial functions that induce the host inflammasome (11-13). Another SPI-7 product, the virulence-associated (Vi) capsular polysaccharide, appears to prevent complement-mediated phagocytosis (14-16). Active immune evasion during epithelium invasion may thus contribute to ‘silent’ crossing of the intestinal barrier, permitting stealthy establishment of systemic infection.
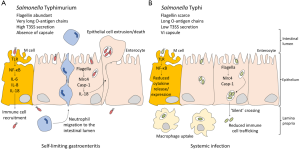
Electron microscopy observation indicated that, unlike S. Typhimurium, S. Typhi invades more frequently enterocytes rather than M cells (7). Microvilli destruction, cell cytoskeleton remodeling and vesicle formation was observed both in biopsy tissues and organoid-derived epithelium monolayers upon S. Typhi infection, but cell death was not detected (7). Transcriptomic analysis revealed downregulation of genes involved in cytoskeletal reorganization and in pathways that control formation of cellular protrusions, cytoplasm organization and microtubule dynamics, among others. Altogether, these observations agree with a previous study that compared S. Typhimurium and S. Typhi interactions with epithelial cells: S. Typhi was found to be less adherent, less invasive and less cytotoxic (17).
Distinct gene expression patterns of Salmonella Typhi and Salmonella Typhimurium during infection
Analysis of S. Typhi gene expression patterns during infection of human biopsies showed discrepancies with S. Typhimurium (7). For instance, genetic loci required for S. Typhimurium invasion like rpoS, ompR and SPI-1 were found to be downregulated in S. Typhi. In contrast, metabolism and ribosomal genes were upregulated during S. Typhi invasion. Upregulation of pathogenicity island SPI-7 during biopsy invasion is consistent, as discussed above, with the role of SPI-7 products in immune evasion (7).
Discrepancies in gene expression patterns between S. Typhi and S. Typhimurium are not surprising: after their evolutionary divergence, each serovar acquired distinct virulence determinants, and S. Typhi underwent genome degradation, a distinctive trait of host-restricted serovars (18). Furthermore, differences between S. Typhi and S. Typhimurium pathogenicity determinants have been previously described. Examples include the structure of the outer core of the LPS (19), the responses to bile (20), and the regulation of flagella and chemotaxis (20).
The model is the message
Animal models are precious tools in translational research if their predictive value to human disease is high enough (21). In the case of typhoid fever, infection of immunodeficient mice by Salmonella Typhimurium has been used for decades to simulate human infection by S. Typhi. Aside from permitting the study of Salmonella interaction with the animal host without the constraints and the dangers of working with a highly pathogenic species, the mouse model has provided a wealth of information on Salmonella virulence mechanisms. However, the steadily growing list of discrepancies between S. Typhimurium and S. Typhi infection mechanisms illustrates the limitations of the immunodeficient mouse model (6) (Figure 1). In medical practice, the limited efficacy of existing typhoid fever vaccines further illustrates such shortages (22). In this scenario, breakthroughs may require the introduction of novel models like the human biopsy cultures and organoids used in the study discussed in this editorial (7). Organoids may be especially appropriate to study infection, immunity, and inflammation as they provide a source of human tissue that accurately reflects human responses (23).
Acknowledgements
We are grateful to Ángela Mérida-Floriano for critical reading of the manuscript.
Funding: Research in our laboratory is supported by grants PCIN-2015-131 (Infect-ERA) and BIO2016-75235-P from the Spanish Ministerio de Economía y Competitividad (MINECO) and the European Regional Fund.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Brenner FW, Villar RG, Angulo FJ, et al. Salmonella nomenclature. J Clin Microbiol 2000;38:2465-7. [PubMed]
- Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol 2014;68:317-36. [Crossref] [PubMed]
- Gunn JS, Marshall JM, Baker S, et al. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 2014;22:648-55. [Crossref] [PubMed]
- Urdaneta V, Casadesús J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med (Lausanne) 2017;4:163. [Crossref] [PubMed]
- Tsolis RM, Xavier MN, Santos RL, et al. How to become a top model: Impact of animal experimentation on human Salmonella disease research. Infect Immun 2011;79:1806-14. [Crossref] [PubMed]
- Johnson R, Mylona E, Frankel G. Typhoidal Salmonella: distinctive virulence factors and pathogenesis. Cell Microbiol 2018;20:e12939. [Crossref] [PubMed]
- Nickerson KP, Senger S, Zhang Y, et al. Salmonella Typhi colonization provokes extensive transcriptional changes aimed at evading host mucosal immune defense during early infection of human intestinal tissue. EBioMedicine 2018;31:92-109. [Crossref] [PubMed]
- Wilson SS, Tocchi A, Holly MK, et al. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol 2015;8:352-61. [Crossref] [PubMed]
- Forbester JL, Goulding D, Vallier L, et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun 2015;83:2926-34. [Crossref] [PubMed]
- Langerholc T, Maragkoudakis PA, Wollgast J, et al. Novel and established intestinal cell line models - An indispensable tool in food science and nutrition. Trends Food Sci Technol 2011;22:S11-20. [Crossref]
- Winter SE, Winter MG, Poon V, et al. Salmonella enterica serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog 2014;10:e1004207. [Crossref] [PubMed]
- Winter SE, Raffatellu M, Wilson PR, et al. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol 2008;10:247-61. [PubMed]
- Winter SE, Winter MG, Atluri V, et al. The flagellar regulator TviA reduces pyroptosis by Salmonella enterica serovar Typhi. Infect Immun 2015;83:1546-55. [Crossref] [PubMed]
- Wilson RP, Winter SE, Spees AM, et al. The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infect Immun 2011;79:830-7. [Crossref] [PubMed]
- Crawford RW, Wangdi T, Spees AM, et al. Loss of very-long O-antigen chains optimizes capsule-mediated immune evasion by Salmonella enterica serovar Typhi. MBio 2013;4:1-8. [Crossref] [PubMed]
- Wangdi T, Lee CY, Spees AM, et al. The Vi capsular polysaccharide enables Salmonella enterica serovar Typhi to evade microbe-guided neutrophil chemotaxis. PLoS Pathog 2014;10:e1004306. [Crossref] [PubMed]
- Bishop A, House D, Perkins T, et al. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: Roles of surface structures in adhesion and invasion. Microbiology 2008;154:1914-26. [Crossref] [PubMed]
- Barquist L, Langridge GC, Turner DJ, et al. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res 2013;41:4549-64. [Crossref] [PubMed]
- Bravo D, Hoare A, Silipo A, et al. Different sugar residues of the lipopolysaccharide outer core are required for early interactions of Salmonella enterica serovars Typhi and Typhimurium with epithelial cells. Microb Pathog 2011;50:70-80. [Crossref] [PubMed]
- Johnson R, Ravenhall M, Pickard D, et al. Comparison of Salmonella enterica serovars Typhi and Typhimurium reveals typhoidal serovar-specific responses to bile. Infect Immun 2018;86:1-16. [PubMed]
- Denayer T, Stöhrn T, Van Roy M. Animal models in translational medicine: Validation and prediction. New Horizons Transl Med 2014;2:5-11.
- Higginson EE, Simon R, Tennant SM. Animal models for salmonellosis: Applications in vaccine research. Clin Vaccine Immunol 2016;23:746-56. [Crossref] [PubMed]
- Iakobachvili N, Peters PJ. Humans in a dish: The potential of organoids in modeling immunity and infectious diseases. Front Microbiol 2017;8:2402. [Crossref] [PubMed]


