Detecting insertion, substitution, and deletion errors in radiology reports using neural sequence-to-sequence models
Introduction
Errors in grammar, spelling, and usage in radiology reports are common. Studies performed between 2008 and 2012 have found errors in 23–36% of final reports for various types of non-plain film imaging (1-5). More recent studies performed with state-of-the-art dictation systems also demonstrate high error rates. A 2015 review of 213,977 radiology reports at the Mayo Clinic found a 9.7% error rate, ranging from a high of a 19.7% in neuroradiology to a low of 3.2% in chest plain films (6). Most significantly, nearly 20% of these reports containing errors were found to contain a clinically material error (6). A 2015 study at a hospital in Staffordshire, UK found that 23% of their CT reports and 32% of their MRI reports contained an error (7). Frequent interruptions and an increasingly fast pace of work contribute to such errors, as well as the widespread adoption of automatic speech recognition systems (8-13). Such systems have the benefit of making reports immediately available after a radiologist has completed dictating, but increase their proofreading responsibility and the amount of time radiologists spend creating reports (14-16).
The use of structured report templates has not been shown to reduce rates of error (3,7). Radiologists believe such errors are far less frequent than actual rates, and such errors may become increasingly problematic in an era when patients can directly see their imaging reports in online patient portals (4,17). The American College of Radiology’s official ‘Practice Parameter for Communication of Diagnostic Imaging Findings’ holds radiologists ultimately responsible for proofreading dictated reports (18). However, many radiologists feel that they do not have adequate time to do so, as indicated in the response of a radiological journal editor in a letter on the topic: “Proofreading of our radiologic reports is required, although we all realize that most of us have limited or no time to do it” (19).
While popular commercially available dictation systems include spell checkers, they do not include systems to detect erroneous word insertions, deletions, or substitutions. Recent advancements in deep learning may be able to support radiologists by checking their reports for such errors, flagging potential errors and suggesting corrections. Ideally, such a system would suggest the precise correction necessary. However, even a system that simply identified sentences likely to require correction could be valuable, as such sentences could be flagged for review before a report was submitted.
Methods
Dataset and pre-processing
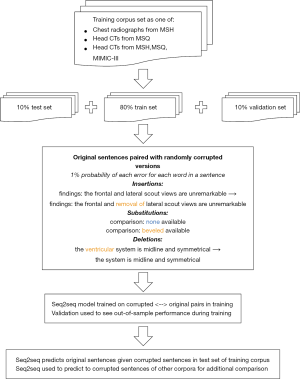
A total of 91,867 head CT and 1,013,287 chest radiograph reports from the Mount Sinai Health System covering a period of 2006–2017 were obtained (Figure 1). These reports were drawn from two sites: Mount Sinai Hospital (MSH) (n=61,722 head CT, 818,978 chest radiograph) and Mount Sinai Queens (MSQ) (n=30,145 head CT,194,309 chest radiograph). These hospitals do not share radiology reporting templates, and are staffed separately by attendings and residents at MSH, and by attendings only at MSQ. A total of 32,259 head CT and 54,685 chest radiograph reports from Beth Israel Deaconess were additionally obtained from the MIMIC-III database (20).
Preprocessing was applied to convert text to lowercase, to convert numbers, times, and numeric dates to a special common token, and to remove all punctuation except for periods, commas, colons, semicolons, and apostrophes. Phrases with both letters and numbers, such as “C3”, which were preserved. Sentences longer than 50 words were truncated; such sentences were extremely uncommon, occurring in only 5,165 of 12,696,846 sentences (0.04%).
Reports were then tokenized into individual sentences. Words that appeared fewer than 10 times in the training corpus were replaced with an ‘unknown’ placeholder token for model training and prediction. As a post-processing step, ‘unknown’ tokens were replaced with the words that mapped to ‘unknown’ tokens at the same position in the corrupted sentence as the ‘unknown’ token (21).
To generate training examples, corruptions of insertion, substitution, and deletion were introduced, each with probability 1% for each word in an original sentence (21). Insertions were drawn from other sentences in the corpus to simulate dictating into the wrong section of a report and ranged between 1–4 words with equal probability. Substitutions consisted of swapping a given word for another word in the report corpus (e.g., “no CT evidence” → substitute ‘hemorrhage’ for ‘CT’ → “no hemorrhage evidence”). Deletions removed a given word from the sentence (e.g., “no CT evidence” → ‘CT’ deleted → “no evidence”). This yielded a set of corrupted sentences, with each matched to an uncorrupted ground truth sentence. This process was repeated 5 times for head CT reports in training, twice for chest radiograph reports in training, and once for both tune and test data. The corrupted sentences served as the input for our model, while the original sentences served as the output.
Modelling approach
Neural sequence-to-sequence (seq2seq) models were then trained to predict ground truth sentences from the corrupted sentences. Seq2seq is a state-of-the-art deep learning architecture that maps an input sequence to an output sequence (22). It has been applied in a variety of natural language processing applications, including machine translation, autoreply, and error correction (21,23-25). This model represents each word in the corpus as a vector of arbitrary length, and subsequently learns a mapping between them. A simple example is presented in Figure 2.
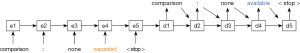
The approach used in this paper adds several features to the basic model of Figure 2 to maximize its predictive performance. It incorporates long short-term memory (LSTM) cells to encode its hidden states, which facilitate learning a more expressive hidden representation (26). It also uses two stacked hidden layers to increase its flexibility. Our seq2seq models are bidirectional to take advantage of the fact that words both preceding and following a given word provide information about its meaning. We also include an ‘attention’ mechanism which allows the model to “attend” to more influential features using the learned attention weights (23).
A PyTorch implementation of seq2seq from OpenNMT-py was used to implement these bidirectional neural seq2seq models, each with 512 hidden units, two layers, and an attention mechanism following Luong (27,28). Word vector embeddings were set to a length of 500. Models were trained with a batch size of 64 on a single Nvidia GTX 1080ti GPU until there was no further improvement in validation perplexity over 3 epochs; this occurred after 5 epochs for single-site chest radiography and 12 epochs for single- and multiple-site head CT. We trained our model utilizing stochastic gradient descent (SGD) with an initial learning rate set to 1.0. This rate was decayed by 50% after any epoch in which perplexity did not decrease on the validation set compared to the prior epoch. The LSTM cells were regularized utilizing dropout at a rate of 0.3, and gradient normalization at a threshold of 5.
Assessment
We were primarily interested in identifying sentences that contained any of these randomly introduced errors. We considered a sentence marked for change when seq2seq predicted a sentence different from the entered input sentence. We were secondarily interested in obtaining a corrected version of that sentence. An accurate correction was defined as a perfect match with the original uncorrupted sentence.
Analysis
Radiology reports were divided into 80% training data, 10% validation data, and 10% test data; all sentences in a given report were included in the same subset. Three separate models were then trained using sentences from the following datasets: (I) head CTs from MSH (n=936,392, 117,716, and 119,412); (II) chest radiographs from MSH (n=6,331,802, 786,779, and 791,217); (III) head CTs from MSH, MSQ, and MIMIC-III (n=2,043,696, 209,030, and 214,169).
Single-modality (head CT or chest radiograph only) model performance was assessed on test cases (I) for the same modality across sites and (II) across modalities at MSH. The jointly trained head CT model (III) was assessed on test cases at each site. For sentences in each test corpus, we report the sensitivity and specificity of the algorithm for identifying sentences with introduced errors. For each group of error (insertion, substitution, deletion, all errors), we also report the percent accurately identified as requiring correction and the percent of original sentences seq2seq exactly recovered for the same-site, same-modality comparisons.
We performed an additional experiment to evaluate the ability of a model trained to synthetic error data to detect real-world radiologist errors in syntax, usage, or spelling in final reports. The MSH head CT model was used to make predictions on uncorrupted test sentences (n=86,211). A sample of 400 flagged sentences and 800 unflagged sentences were manually reviewed by one of the authors (JZ). He was blinded to the algorithm’s predictions and labeled these sentences for any kind of apparent error in usage, syntax, or spelling. Positive predictive value (PPV) and negative predictive value (NPV) were calculated for this sample. To facilitate comparison with sensitivity and specificity results from other experiments, the PPV and NPV calculated for this sample were used to estimate sensitivity and specificity for the full group of n=86,211 uncorrupted sentences. This was done by randomly drawing PPV* and NPV* from their respective posterior distributions under a uniform prior [i.e., beta(1+correct_cases,1+incorrect_cases)], assigning all seq2seq-changed and unchanged cases positive for a true error with probability PPV* and 1-NPV*, respectively, calculating sensitivity and specificity for the full n=86,211 sentences, and repeating this process 10,000 times to generate a 95% confidence interval.
Results
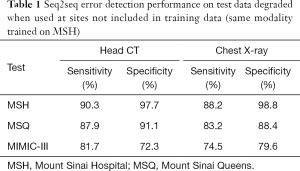
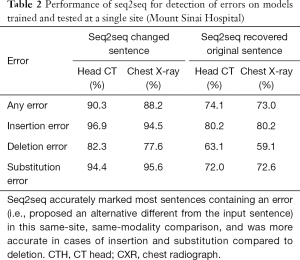
In same-site, same-modality test sets, seq2seq detected 90.3% of corrupted sentences in head CTs and 88.2% in chest radiographs with 97.7% and 98.8% specificity, respectively (Table 1). For errors introduced in same-site, same-modality test sets, seq2seq recovered 74.1% of the original head CT sentences and 73.0% of the chest radiograph sentences (Table 2). When it did not recover the original sentence, it sometimes offered reasonable alternatives. Examples from the same-site head CT test set are given in Figure 3.
Full table
Full table
Seq2seq performance degraded when test sites (Table 1) and modalities (Table 3) were not included in training but performed well when trained jointly on multiple sites (Table 4). Performance was strongest in the same-site, same-modality comparison as radiology reports are typically standardized within a site but vary between sites, and the language of reports is highly specific to modality. For head CT, sensitivity decreased from 90.3% to 87.9% and 81.7% in two external test sets, and specificity decreased from 97.7% to 91.1% and 72.3%. For chest radiography, sensitivity decreased from 88.2% to 83.2% and 74.5% in these external test sets, and specificity decreased from 98.8% to 88.4% and 79.6%. Performance decreased sharply when the head CT model was used to identify errors in same-site chest radiography reports (sensitivity 70.0%, specificity 81.1%) and vice-versa (sensitivity 70.0%, specificity 72.8%).
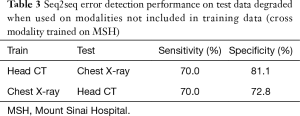
Full table
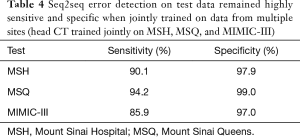
Full table
A seq2seq model jointly trained on head CT reports from all three collections achieved a sensitivity of 90.1%, 94.2%, and 85.9% and specificity of 97.9%, 99.0%, and 97.0% at each site, respectively.
Manual review of 400 seq2seq-flagged and 800 unflagged same-site same-modality original uncorrupted head CT sentences to estimate real-world performance demonstrated errors in usage, syntax, or spelling in 157/400 seq2seq-flagged (PPV 0.393, 95% CI, 0.346–0.441) and 11/800 unflagged (NPV 0.986, 95% CI, 0.976–0.992) sentences. Estimated sensitivity over the n=86,211 unmodified training examples was 0.389 (95% CI, 0.267–0.542) with specificity 0.986 (95% CI, 0.985–0.987). Examples of real-world errors are included in Table 5.
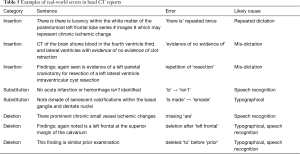
Full table
Discussion
When trained on reports for a specific modality at a specific site, seq2seq can effectively detect random insertion, deletion, and substitution errors introduced into radiology reports (sensitivity 90.3% and 88.2%, specificity 97.7% and 98.8% for head CT and chest radiograph, respectively). While accurate prediction is desirable, we note that we do not require accurate predictions for seq2seq to function well in its primary role of detecting errors, where it achieved high sensitivity and specificity. In the majority of cases (74.1% and 73.0%), it reproduced the exact sentence from the original report. This performance yields a conservative lower bound on the actual accuracy, as synonymous but different phrases (e.g., “I agree with the resident’s interpretation” vs. “I agree with the resident’s interpretation thereof”) would not be counted as a correct recovery. Seq2seq performed best at detecting inappropriate insertions (96.9% and 94.5%, respectively, for head CT and chest radiographs) and substitutions (94.4% and 95.6%, respectively) compared to deletions (82.3% and 77.6%, respectively). Intuitively, insertions and substitutions introduce out-of-place content into a sentence and typically stand out, whereas deletions can be subtler (e.g., deleting an adjective).
Seq2seq can learn detailed modality-specific and site-specific patterns, which is facilitated by the limited lexical complexity of radiology corpora, but these models have limited generalization (29). Predictions for head CT and chest radiography reports had substantially lower specificity when used to detect errors at a new site or on a new modality. High specificity is critical to the practical utility of a proofreading tool for radiology reports: one cannot reasonably imagine that radiologists under time pressure will elect to use a tool that requires them to sift through many false positives. Accordingly, we believe these models would need to be trained with data from individual sites to achieve maximal specificity. Our results suggest that such a model could be jointly trained on data from multiple sites while maintaining predictive accuracy, as a head CT model jointly trained on data from all 3 sites had excellent predictive accuracy at all three. While site-specific data would need to be provided, the proofreading engine itself could be deployed as a software-as-a-service tool requiring no engineering on the user side.
Future work could explore how the probability of sentence corruption can function as a tuning parameter. Because of the randomness of this corruption process, a mix of uncorrupted and corrupted sentences were included in train and test data. For example, in MSH test data, 27.8% of head CT and 24.8% of chest radiograph sentences were corrupted. This prevented the model from trivially learning to change every sentence and determined seq2seq’s sensitivity and specificity threshold.
Our approach is limited by the fact that it considers each sentence of a report in isolation, eliminating the possibility of detecting inconsistencies between different sections of a report (e.g., “unremarkable study” in Impression should only appear if there is no description of any acute pathology in Findings). Other work has shown how seq2seq models can be extended hierarchically to encode paragraphs from building blocks of words and sentences, and our approach could extend to the level of a full report (30). Additional training data would likely be required to provide sufficient training examples for a hierarchical seq2seq model.
Our models are also limited by our use of simulated error data. While this presents a unique opportunity for creating training data, it is not ideal: some of our ground truth data contains uncorrected errors, as we demonstrated in our manual review. A stronger dataset would consist of radiology reports containing errors and corresponding manual corrections, but the scale at which seq2seq requires training data makes such an approach prohibitively expensive. Numerous reports would have to be reviewed to find sufficient examples of errors because an overwhelming number of radiology reports are error-free. For example, based on our manual review, we estimate that sentences in the MSH head CT corpus contain an error in 2.2% of cases. Because reports contain many sentences, the error rate per sentence is substantially lower than the error rate per report. The vast majority of original sentences used as ground truth data are therefore free of errors and can serve as a useful basis on which to simulate errors.
We believe further refinement is needed to improve performance before such a model will be able to detect all types of real-world errors. The MSH head CT model demonstrated an estimated same-site sensitivity of 0.389 (95% CI, 0.267–0.542) and specificity of 0.986 (95% CI, 0.985–0.987) in detecting real-world errors in reports. This demonstrates the potential of this approach to detect real-world errors that made it into final reports despite being trained exclusively on randomly generated errors. We note that this estimation is limited by its dependence on a very small number of observed false negatives (n=11) in our manually reviewed data, which is responsible for the observed wide confidence interval over sensitivity. Real-world errors can be much subtler than the ones randomly introduced into our training data; for example, one false negative case was deemed incorrect because a plural verb was used with a singular subject (“Vascular calcification is also quite prominent and quite distal which likely reflect severe disease”), while another was missing “of” (“No evidence large infarct, parenchymal hemorrhage, or mass effect”). The inclusion of additional simulated errors, especially those most likely to adversely affect patient care (adding/removing negation, inverting laterality, etc.), would likely improve performance. It may also be possible to incorporate additional training data from radiologists themselves. Attending radiologists routinely review and correct draft resident reports, and many systems track changes made; these could be included as training data. With collaborating radiologists and necessary technical infrastructure, it would be possible to track radiologist report edits in real-time and learn from these examples how to correct common types of mistakes. We believe iterative improvements such as these could improve seq2seq’s sensitivity and specificity.
Conclusions
Seq2seq models can be highly effective at detecting erroneous insertions, deletions, and substitutions of words in radiology reports. To achieve high performance, these models require site- and modality-specific training examples. Incorporating additional targeted training data can further improve performance in detecting real-world errors in reports.
Acknowledgments
We would like to thank Burton Dreyer, MD, for his support of this work.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Mount Sinai Health System Institutional Review Board. The requirement for patient consent was waived for this retrospective study that was deemed to carry minimal risk. All data was stored exclusively on a secured, dedicated HIPAA-compliant computing resource on the Mount Sinai Hospital campus.
References
- Chang CA, Strahan R, Jolley D. Non-clinical errors using voice recognition dictation software for radiology reports: a retrospective audit. J Digit Imaging 2011;24:724-8. [Crossref] [PubMed]
- Pezzullo JA, Tung GA, Rogg JM, et al. Voice recognition dictation: radiologist as transcriptionist. J Digit Imaging 2008;21:384-9. [Crossref] [PubMed]
- Hawkins CM, Hall S, Hardin J, et al. Prepopulated radiology report templates: a prospective analysis of error rate and turnaround time. J Digit Imaging 2012;25:504-11. [Crossref] [PubMed]
- Quint LE, Quint DJ, Myles JD. Frequency and spectrum of errors in final radiology reports generated with automatic speech recognition technology. J Am Coll Radiol 2008;5:1196-9. [Crossref] [PubMed]
- Basma S, Lord B, Jacks LM, et al. Error rates in breast imaging reports: comparison of automatic speech recognition and dictation transcription. AJR Am J Roentgenol 2011;197:923-7. [Crossref] [PubMed]
- Ringler MD, Goss BC, Bartholmai BJ. Syntactic and semantic errors in radiology reports associated with speech recognition software. Health Informatics J 2017;23:3-13. [Crossref] [PubMed]
- Najran P. Errors using voice recognition software in radiology reports. Poster presented at: European Congress of Radiology 2015 March 4-8, Vienna, Austria.
- Williams LH, Drew T. Distraction in diagnostic radiology: How is search through volumetric medical images affected by interruptions? Cogn Res Princ Implic 2017;2:12. [Crossref] [PubMed]
- Yu J-PJ, Kansagra AP, Mongan J. The radiologist’s workflow environment: evaluation of disruptors and potential implications. J Am Coll Radiol 2014;11:589-93. [Crossref] [PubMed]
- Ellenbogen PH. The "p word". J Am Coll Radiol 2012;9:603. [Crossref] [PubMed]
- Chokshi FH, Hughes DR, Wang JM, et al. Diagnostic Radiology Resident and Fellow Workloads: A 12-Year Longitudinal Trend Analysis Using National Medicare Aggregate Claims Data. J Am Coll Radiol 2015;12:664-9. [Crossref] [PubMed]
- Strahan RH, Schneider-Kolsky ME. Voice recognition versus transcriptionist: error rates and productivity in MRI reporting. J Med Imaging Radiat Oncol 2010;54:411-4. [Crossref] [PubMed]
- du Toit J, Hattingh R, Pitcher R. The accuracy of radiology speech recognition reports in a multilingual South African teaching hospital. BMC Med Imaging 2015;15:8. [Crossref] [PubMed]
- Hammana I, Lepanto L, Poder T, et al. Speech recognition in the radiology department: a systematic review. Health Inf Manag 2015;44:4-10. [PubMed]
- Prevedello LM, Ledbetter S, Farkas C, et al. Implementation of speech recognition in a community-based radiology practice: effect on report turnaround times. J Am Coll Radiol 2014;11:402-6. [Crossref] [PubMed]
- Williams DR, Kori SK, Williams B, et al. Journal Club: Voice recognition dictation: analysis of report volume and use of the send-to-editor function. AJR Am J Roentgenol 2013;201:1069-74. [Crossref] [PubMed]
- Bruno MA, Petscavage-Thomas JM, Mohr MJ, et al. The “Open Letter”: Radiologists’ Reports in the Era of Patient Web Portals. J Am Coll Radiol 2014;11:863-7. [Crossref] [PubMed]
- American College of Radiology. ACR Practice Parameter for Communication of Diagnostic Imaging Findings, 2014.
- Berlin L. Liability for typographical errors. AJR Am J Roentgenol 2011;196:W215. [Crossref] [PubMed]
- Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016;3:160035. [Crossref] [PubMed]
- Paino A. deep-text-corrector [Accessed September 18, 2017]. Available online: https://github.com/atpaino/deep-text-corrector
- Sutskever I, Vinyals O, Le QV. Sequence to Sequence Learning with Neural Networks. In: Ghahramani Z, Welling M, Cortes C, et al. editors. Advances in Neural Information Processing Systems 2014, 8-13 December 2014, Montreal, Canada. p. 3104-12.
- Bahdanau D, Cho K, Bengio Y. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv [cs.CL] 2014. Available online: http://arxiv.org/abs/1409.0473
- Kannan A, Kurach K, Ravi S, et al. Smart Reply: Automated Response Suggestion for Email. arXiv [cs.CL] 2016. Available online: https://arxiv.org/abs/1606.04870
- Sak H, Senior A, Beaufays F. Long Short-Term Memory Recurrent Neural Network Architectures for Large Scale Acoustic Modeling. Paper presented at: INTERSPEECH 2014: Fifteenth Annual Conference of the International Speech Communication Association, 14-18 September 2014, Singapore. p. 338-42.
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. [Crossref] [PubMed]
- Klein G, Kim Y, Deng Y, et al. OpenNMT: Open-Source Toolkit for Neural Machine Translation. In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics-System Demonstrations, 30 July-4 August 2017, Vancouver, Canada. p. 67-72.
- Luong M-T, Pham H, Manning CD. Effective Approaches to Attention-based Neural Machine Translation. arXiv [cs.CL] 2015. Available online: http://arxiv.org/abs/1508.04025
- Zech J, Pain M, Titano J, et al. Natural language based machine learning models for the annotation of clinical radiology reports. Radiology 2018;287:570-80. [Crossref] [PubMed]
- Li J, Luong M-T, Jurafsky D. A Hierarchical Neural Autoencoder for Paragraphs and Documents. arXiv [cs.CL] 2015. Available online: http://arxiv.org/abs/1506.01057