Two novel mutations in parE among Shigella flexneri isolated from Jiangsu Province of China, 2016
Introduction
With the frequent mobility of international population, bacillary dysentery (or called shigellosis), a severe intestinal infection disease caused by Shigella spp, has become a great threat on human’s health (1,2). The endemic epidemic and widespread of pathogens have exposed the children and the elderly to be the primary targets of the diarrheal disease, especially for those in the areas with relatively poor economy and unqualified sanitation. People in these areas are more likely to develop bad living habits. As a result, they could easily get in touch with the infectious bacteria involved in the contaminative water and food (3-5). In China, Shigella has already been listed into the Chinese Center for Disease Control and Prevention as one of the most important pathogenic bacteria of gastrointestinal infection since 2006, and it calls for more attention and further development (6).
Furthermore, by the classification based on O antigen, Shigella consisted of 4 different subgroups, S. dysenteriae, S. flexneri, S. boydii, and S. sonnei. Among some developed and (or) industrialized countries that were reported before, S. sonnei has not been limited to the main emergence yet, but it gradually increases the proportion in some regions with notable shift of socio-economic types (5,7,8). Even so, it was reported that the diverse S. flexneri, with 15 serotypes or subtypes, remains the most prevalent pathogenic factor leading to the gastrointestinal disease in many developing countries (9).
A lot of antibacterial agents, especially the ciprofloxacin, have been advised as the first-line antibiotics against the serious Shigella infections. These antibacterial agents have been widely applied into the clinical treatment for shigellosis in the past years (10,11). However, with the sharp development of fluoroquinolone resistance or the more severe multiple-drug resistance in Asia and European regions (4,12,13), it’s actually a tremendous challenge to pick out the most appropriate antibiotic. In the previous studies, many target mutations of quinolone resistance-determining region (QRDR) have been reported. The mutations in gyrA and parC may be responsible for the fluoroquinolone-resistant S. flexneri as the primary mechanism (14,15). Besides the high prevalence of amino acid alterations in gyrA, gyrB and parC, no one can show that there are other mutations in parE gene within S. flexneri isolates. What’s more, plasmid-mediated quinolone resistance (PMQR) genes were associated with mobile elements. It also plays an important role on the poor resistance capacity against fluoroquinolones, leading to the increase of the minimum inhibitory concentration (MIC) and the loss of original drug activity (15-17). Besides, as Liu and colleagues have reported in 2012, multiple-resistant clinical isolates of S. flexneri were correlated to the influence of some resistance determinants, such as integrons and β-lactamases (18).
We have successively carried out detailed characterization and studies on S. flexneri isolates in Jiangsu Province of China from 2001 to 2015, especially in the aspect of antimicrobial resistance to quinolones and relevant molecular mechanism. The results of these studies have displayed the essentiality of continuous monitoring in this area (19,20). Based on this, we collected the S. flexneri isolates in Jiangsu Province of China in 2016, trying to observe the changes of resistance to fluoroquinolones, the prevalence of PMQR determinants, and ulteriorly explore whether new point mutation associated with resistance mechanism exists in the QRDR genes or not.
Methods
Bacterial collection
With the great support of Jiangsu Provincial Center for Disease Control and Prevention, 81 strains of S. flexneri in Jiangsu Province of China during 2016 were collected. All these samples were from clinical patients of 12 cities, including Southern Jiangsu (n=27), Central Jiangsu (n=18) and Northern Jiangsu (n=36) in this area. By the way, Yangzhou was not included.
Strains identification and serotyping
After the recovery of strains, automatic VITEK2 COMPACT analysis system was applied into the bacterial identification under the guidance of the manufacturer (BioMerieux, Marcy l’ Etoile, France). And serotyping of 81 isolates was confirmed by slide agglutination in combination with specific antisera of Shigella species (Tianrun Bio-Pharmaceutical Co., Ltd., China).
Susceptibility test in quinolones
Based on the disc diffusion method (Kirby-Bauer), M-H and N-A agar (Oxoid, Hampshire, UK) were employed to test the isolates’ susceptibility to nalidixic acid and ciprofloxacin. Escherichia coli ATCC 25922 and ATCC 35218 were cultured for quality control, and the result of susceptibility test was judged by Clinical and Laboratory Standards Institute (CLSI) standards (21).
Genes amplification and products sequencing
Study on QRDR
PCR was applied to amplify the genes of QRDR, including gyrA, gyrB, parC and parE, using primers synthesised in Sangon Biotech (Shanghai, China). It also referred to the previous studies (22-24). PCR products were verified by agarose gel electrophoresis, and then were sent to Genewiz Company (Suzhou, China) for nucleotide sequencing after being purified. The results were analyzed by the Basic Local Alignment Search Tool (BLAST) comparison with sequences in the GenBank database.
Study on PMQR
Screening of PMQR determinants (qnrA, qnrB, qnrC, qnrD, qnrS, aac(6’)-Ib-cr and qepA) was performed by PCR amplification, based on published primers (17,25-28). Agarose gel electrophoresis was used to observe the genes above. And then, the purified products with positive genes were sequenced in Genewiz Company (Suzhou, China) and compared with the sequences in the GenBank to ensure the existence of PMQR determinants.
Statistical analysis
The software SPSS 19.0 was used for data management and statistical analysis. Comparisons of serotypes distribution, mutations and resistance were judged by P value. Only when P≤0.05, the research was considered to be valuable.
Results
Shigella isolates and serotypes
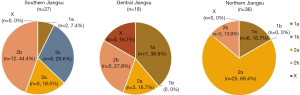
Coming from 12 cities in Jiangsu Province, China during 2016, a total of 81 S. flexneri isolates were sponsored by Jiangsu Provincial Center for Disease Control and Prevention. All these strains were serotyped and 5 different serotypes were identified, among them 2a and 2b being the most common serotypes, accounting for 40.7% (33/81) and 27.2% (22/81) (P<0.05). Nevertheless, 1a, 1b and X only occupied a smaller proportion of 18.5% (15/81), 9.9% (8/81) and 3.7% (3/81). What’s more, the serotypes distribution of Shigella in 3 regions had significant differences (P<0.001). When it comes to the type of serotypes, it’s obvious that Northern Jiangsu obtained a higher proportion of 2a isolates with the percentage of 69.4%, while 2b isolates had a percentage of 44.4% in Southern Jiangsu. Central Jiangsu was different from both of the mentioned regions, for 38.9% proportion was occupied by 1a isolates there. Meanwhile, 3 X S. flexneri strains were all isolated from Taizhou in Central Jiangsu without exception (Table 1, Figure 1).
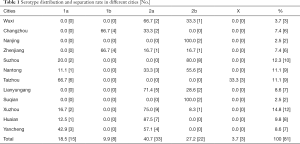
Full table
Quinolone susceptibility test
Resistance and regions
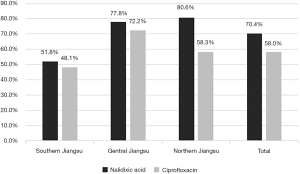
Among these isolates, 70.4% were resistant to nalidixic acid and 58.0% (47/81) expressed resistance to ciprofloxacin. Overall, S. flexneri isolates in the 3 regions of Jiangsu Province conferred obviously different resistant level to nalidixic acid (P<0.05). In particular, the isolates in the region of Northern Jiangsu possessed the highest resistance rate of 80.6%, while the Southern Jiangsu (51.8%). Nevertheless, the ciprofloxacin-resistant strains mainly focused on the area of Central Jiangsu (72.2%), followed by the isolates in Southern Jiangsu (48.1%) and Northern Jiangsu (58.3%) (Figure 2).
Resistance and serotypes
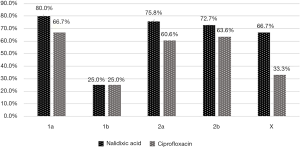
Additionally, in some degree, the resistant level of S. flexneri isolates to quinolones also presented a slight difference between the diverse serotypes. Among the 5 serotypes detected, 1a isolates simultaneously possessed the highest resistance to nalidixic acid and ciprofloxacin, with the percentage of 80.0% and 66.7%, respectively. These two proportions were both a little higher than 2a (75.8% and 60.6%) and 2b (72.7% and 63.6%). The more interesting fact is that 1b S. flexneri expressed the same resistance rate to both 2 antibacterial drugs, accounting for 25.0%, but obviously lower than the other 4 serotypes (P<0.05) (Figure 3).
Target mutations in QRDR genes
Mutations in genes of topoisomerase IV
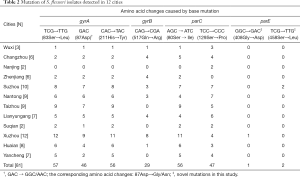
With the methods of sequences comparison, 2 novel mutations in parE were observed from 3 isolates, and 2 of them were detected with mutation at codon 458 (TCG→TTG), contributing to Serine replaced by Leucine. Meanwhile, another mutation presented with the substitution of Gly408Asp (GGC→GAC) within a single isolate. In our opinion, all these 2 mutations in parE had never been reported before, for the gene of parC, 69.1% isolates showed the common alteration of Ser80Ile. So it was the same with did the mutation His211Tyr in gyrA, higher than the other mutation at position of 129 in parC (Ser→Pro) with the percentage of 58.0% (Table 2).
Full table
Mutations in genes of DNA gyrase
Besides, a total of 3 point mutations were detected in gyrA, of which, the amino acid substitution at the position of Ser83 and His211 were the most 2 prevalent, accounting for 70.4% and 69.1%, respectively, followed by the mutation of Asp87Gly/Asn (56.8%). Besides, 35.8% of isolates showed the alteration of Gln517Arg in gyrB, which was much lower than the mutation rate in gyrA and parC (Table 2).
Mutations and resistance
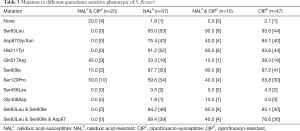
Among 57 nalidixic acid-resistant isolates, mutation Ser83Leu and His211Tyr in gyrA played a very important part, accounting for 93.0% and 91.2%, followed by Ser80Ile within parC, which was obviously higher than other mutations (P<0.05). Additionally, with 3 point mutations, including Ser83Leu (gyrA), Ser80Ile (parC) and Asp87Gly/Asn (gyrA), isolates remained a high prevalence of 76.6% resistance rate to ciprofloxacin. Noticeably, almost half of ciprofloxacin-susceptible and nalidixic acid-susceptible Shigella isolates generated amino acid mutation at the position of Ser129 in parC, followed by the mutation in gyrB (Gln517Arg). For parE, carrying mutation Ser458Leu, 2 isolates didn’t have susceptibility to ciprofloxacin and nalidixic acid any more, while the single isolate with mutation Gly408Asp was resistant to nalidixic acid but susceptible to ciprofloxacin. And we also observed that one strain of Shigella did not have any mutation, but resistant to both 2 drug detected (Table 3).
Full table
Mutations and serotypes
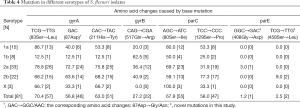
Besides, 86.7% 1a S. flexneri isolates generated the mutation of Ser83Leu, followed by Ser80Ile in parC (80.0%), remarkably higher than other mutations (P<0.05). While serotype 2a isolates presented a more prevalence of gyrA mutation, floating between 72.7% and 78.8%. It was different that serotype 2b isolates mainly carried with the mutation at codon 129 in parC, accounting for 77.3%. Moreover, 2 X serotype strains with mutations of Ser83Leu and His211Tyr in gyrA also generated the substitution in parC at the position of Ser80 synchronously. Contrary to other 4 serotypes, 1b tended to be the lowest mutation rate in all the mutations within the gene of gyrA (P<0.05) (Table 4).
Full table
Analysis of PMQR genes
Among 81 S. flexneri isolates, 6 strains were detected with PMQR positive genes, including 1 aac(6’)-Ib-cr and 6 qnrS positive isolates, without the participation of qnrA, qnrB, qnrC, qnrD and qepA. Every positive isolate involved at least 3 mutations, especially all the 6 qnrS positive isolates mutated at Ser83Leu in gyrA in combination with the alteration of Ser80Ile in gyrC except the number 68. Even 4 of them generated the mutation simultaneously at codon 87 in gyrA. Furthermore, mutations Gln517Arg in gyrB and Ser129Pro in parC were also detected among 2 and 4 positive isolates. Besides, the single aac(6’)-Ib-cr positive strain was observed with the emergency of qnrS gene. Unfortunately, all these PMQR positive isolates showed resistance to nalidixic acid and even 5 of them didn’t keep susceptible to ciprofloxacin any more (Table 5).

Full table
Discussion
Looking back to our research, 5 novel mutations have been detected in S. flexneri isolated from Jiangsu Province of China during 2001–2011, including 2 mutations in gyrA (Asn57Lys and His80Pro) and 3 mutations in parC (Ala85Thr, Asp111His and Ser129Pro) (20). In this thesis, 2 novel mutations in parE of the topoisomerase IV at Ser458Leu and Gly408Asp were identified among 3 isolates. From our perspective, all the 2 mutations had never been reported before.
Meanwhile, susceptibility test result demonstrated that 2 isolates with the mutation
Ser458Leu all expressed resistance to nalidixic acid and ciprofloxacin, while the single isolate harboring the alteration of Gly408Asp was resistant to nalidixic acid but susceptible to ciprofloxacin. The change of amino acid could indirectly affect the combination of enzymes and fluoroquinolones, and then resulted in the emergency of resistance (29). All the 2 novel mutations had amino acid changes, especially the serine at codon 458 in parE was replaced by leucine, so it’s possible that the mutation Ser458Leu could also lead to the same level of fluoroquinolones resistance as Ser83Leu (gyrA) did. What’s more, a previous study proved that the mutation Ser458Ala in parE found from Escherichia coli was in correlation with the increase in MIC for ciprofloxacin (30). Therefore, both the 2 novel mutations in parE of S. flexneri have the potential to increase the MIC for ciprofloxacin and mediate fluoroquinolone resistance. However, it still needs to explore in a further way.
Indeed, this is an interesting result that strains of S. flexneri in the 3 regions of Jiangsu Province had evident differences in serotype distribution. Serotype 2b was mainly reflected in Southern Jiangsu, which was discrepant to Central Jiangsu (serotype 1a) and Northern Jiangsu (serotype 2a). Unbalanced economic status or other social factors may be an explanation for this phenomenon. Obviously, 1a isolates seemed to be the most threatening type (with the highest resistance to both nalidixic acid and ciprofloxacin), revealing that Central Jiangsu should be highly aware of the serious situation of anti-infection, and solve the problem in a more rational way of controlling antibiotics. Meanwhile, they are supposed to bear in mind that human immune specificity, unknown cross protection, and diverse serotypes with distribution difference mean stricter requirements for the development of vaccines in order to prevent the dissemination of pathogenic bacteria (31-33).
In recent years, several regions in the Eastern provinces of China, such as Henan (34), Zhejiang (35) and Anhui (36), have reported a certain level of ciprofloxacin resistance of S. flexneri, ranging from 21.0% to 25.4%, which was much lower than our data (58.0%). Hence, the frequent occurrence of ciprofloxacin resistance requires a further specific test on whether fluoroquinolones can be applied into the treatment or not. Under this pressure, more efforts are supposed to be made to the study on fluoroquinolone resistance mechanism.
Up to now, chromosomal mutation in target genes of QRDR remains the most significant mechanism for fluoroquinolone resistance, particularly the amino acid substitution in gyrA and parC (15,37), and a recent study based on structural level has reconfirmed this (38). In this research, unsurprisingly, Ser83Leu within gyrA was still described as the most dominant point mutation, consistent with previous study (19), with gyrA (His211) and parC (Ser80) being the predominant. Besides, among 57 nalidixic acid-resistant isolates, mutation Ser83Leu and His211Tyr in gyrA also played an important part. It could not be ignored that since the mutation His211Tyr (gyrA) was first detected in Bangladesh during 2009 (39), the quinolone-resistant S. flexneri isolates seemed to get more attention (19,40,41), and this made us keep thinking that whether this mutation could confer resistance to antibacterial agents. Then, our data also showed that multiple mutations will highly increase the risk of acquiring fluoroquinolone resistance (37), for 76.6% Shigella isolates expressed resistance to ciprofloxacin with 3 critical mutations at the same time, which were located at the position of Ser83 and Asp87 within gyrA in company with Ser80 in parC.
However, no evidence could prove that the mutations have to appear in the antimicrobial-resistant strains, so neither the susceptible ones. In this study, almost half of strains showing susceptibility to both nalidixic acid and ciprofloxacin were observed with mutation in parC (Ser129Pro) or gyrB (Gln517Arg), and similar research findings also appeared in Zhejiang, China (35). The chance was that we need to combine the additional mutations in these target genes or determinants associated with plasmid together thus mediating the high level of resistance far from the decreased susceptibility (15,42). Nevertheless, 1 single Shigella isolate was corroborated to be resistant to both 2 drugs tested, but none of mutations occurred, which may be attributed to decreased drug accumulation or other mechanisms (15).
Besides, obvious difference between mutations and serotypes was also observed among 81 S. flexneri isolates. For instance, Ser83Leu in gyrA and Ser80Ile in parC among 1a isolates were more prevalent than other mutations, which may be a reason for the high resistance rate of this serotype to antimicrobial agents mentioned above. However, contrary to other 4 serotypes, the emergency of mutations in gyrA among 1b isolates only takes up a small proportion, indicating that the selection of mutation was associated with serotypes. What’s more, it also certified that serological research was also an effective means to better analyze it.
Since the end of the twentieth Century, qnr families, aac(6’)-Ib-cr and qepA, involved with protection, inactivity and efflux, gradually went into people’s sight and were described as PMQR determinants by participating in low level of fluoroquinolone resistance (16,17,43,44). Here, qnrS and aac(6’)-Ib-cr were observed within six clinical isolates. It was different from our previous result (19) that qnrS became the most prevalent positive gene. In addition, all qnrS positive strains that conferred resistance to ciprofloxacin were involved in at least 3 mutations in gyrA and parC, especially at the position of Ser83 and Ser80. It suggested that qnr family played an important part in protecting the target sites of fluoroquinolone, and in accelerating the emergency of chromosomal mutation. Subsequently, all of these made preparations for the high level of resistance (17). However, 5 of 6 PMQR positive strains lost original susceptibility to ciprofloxacin, which tended to be more serious than other regions in China, such as Anhui (36). Besides, the single aac(6’)-Ib-cr positive isolate was observed in co-existence of qnrS gene, which was in agreement with what Luo et al. has reported before (45). However, it has to be noticed that horizontal transmission of resistance gene could increase the risk of acquiring resistance. Thus, it’s reasonable enough to constantly monitor the dynamic changes of these PMQR genes, and to seasonably develop evasive strategies according to the latest normalized data.
Unavoidably, there were some shortcomings in our study. To begin with, the current samples could only reflect the basic profile of S. flexneri in the year 2016, but could not make systematic review of the trend analysis. In addition, it is still unclear whether the new mutations in parE correlate with resistance, and which will be our next plan. Last but not least, there may be some differences between drug susceptibility test and the result in vivo test, because some regions show us the high drug resistance. Therefore, it is necessary to verify the true resistance combined with the clinical efficacy of patients.
Conclusions
In this study, we have made a specific analysis on the S. flexneri isolates from Jiangsu Province of China in 2016, concentrating on the resistance to quinolone and quinolone resistance mechanism involved. In our thesis, a breakthrough is that 2 novel mutations were identified in parE gene of QRDR. Apart from this, the resistance of S. flexneri isolates to fluoroquinolone was documented to be extremely high. Besides, given that the emergency of PMQR determinants, it’s high time to strengthen the communication between laboratory and clinical. We should also make out effective strategies accordingly in order to control the spread of pathogenic bacteria and reduce the burden of shigellosis.
Acknowledgements
Funding: This work was supported by the Jiangsu Students’ Platform for innovation and entrepreneurship training program (201710313075X), the Chinese National Natural Science Foundation (81471994, 81702103), the Natural Science Foundation of Jiangsu Province (BK20151154), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six talent peaks project of Jiangsu Province (WSN-135), and the Advanced Health Talents of Six-one Project of Jiangsu Province (LGY2016042).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Folster JP, Pecic G, Bowen A, et al. Decreased susceptibility to ciprofloxacin among Shigella isolates in the United States, 2006 to 2009. Antimicrob Agents Chemother 2011;55:1758-60. [Crossref] [PubMed]
- De Lappe N, O'Connor J, Garvey P, et al. Ciprofloxacin-Resistant Shigella sonnei Associated with Travel to India. Emerg Infect Dis 2015;21:894-6. [Crossref] [PubMed]
- WHO. Guidelines for the control of epidemics due to Shigella dysenteriae type 1. World Health Organization, 2005. Available online: http://www.who.int/topics/cholera/publications/WHO_CDR_95_4/en/
- von Seidlein L, Kim DR, Ali M, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med 2006;3. [Crossref] [PubMed]
- Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 1999;77:651-66. [PubMed]
- Zhang J, Wang F, Jin H, et al. Laboratory monitoring of bacterial gastroenteric pathogens Salmonella and Shigella in Shanghai, China 2006-2012. Epidemiol Infect 2015;143:478-85. [Crossref] [PubMed]
- Zhang H, Si Y, Wang X, et al. Patterns of Bacillary Dysentery in China, 2005-2010. Int J Environ Res Public Health 2016;13:164. [Crossref] [PubMed]
- Holt KE, Baker S, Weill FX, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet 2012;44:1056-9. [Crossref] [PubMed]
- Qiu S, Xu X, Yang C, et al. Shift in serotype distribution of Shigella species in China, 2003-2013. Clin Microbiol Infect 2015;21:252.e5-8. [Crossref] [PubMed]
- Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. Available online: http://www.who.int/cholera/publications/shigellosis/en/
- Acar JF, Goldstein FW. Trends in bacterial resistance to fluoroquinolones. Clin Infect Dis 1997;24 Suppl 1:S67-73. [Crossref] [PubMed]
- Gu B, Cao Y, Pan S, et al. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents 2012;40:9-17. [Crossref] [PubMed]
- Nüesch-Inderbinen M, Heini N, Zurfluh K, et al. Shigella Antimicrobial Drug Resistance Mechanisms, 2004-2014. Emerg Infect Dis 2016;22:1083-5. [Crossref] [PubMed]
- Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 2005;25:358-73. [Crossref] [PubMed]
- Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 2003;51:1109-17. [Crossref] [PubMed]
- Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet 1998;351:797-9. [Crossref] [PubMed]
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 2006;6:629-40. [Crossref] [PubMed]
- Liu Y, Hu L, Pan Y, et al. Prevalence of plasmid-mediated quinolone resistance determinants in association with β-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J Med Microbiol 2012;61:1174-6. [Crossref] [PubMed]
- Qin T, Qian H, Fan W, et al. Newest data on fluoroquinolone resistance mechanism of Shigella flexneri isolates in Jiangsu Province of China. Antimicrob Resist Infect Control 2017;6:97. [Crossref] [PubMed]
- Qin T, Bi R, Fan W, et al. Novel mutations in quinolone resistance-determining regions of gyrA, gyrB, parC and parE in Shigella flexneri clinical isolates from eastern Chinese populations between 2001 and 2011. Eur J Clin Microbiol Infect Dis 2016;35:2037-45. [Crossref] [PubMed]
- Performance standards for antimicrobial susceptibility testing: twenty-third informational supplement. Clinical and Laboratory Standards Institute, 2013.
- Rahman M, Mauff G, Levy J, et al. Detection of 4-quinolone resistance mutation in gyrA gene of Shigella dysenteriae type 1 by PCR. Antimicrob Agents Chemother 1994;38:2488-91. [Crossref] [PubMed]
- Dutta S, Kawamura Y, Ezaki T, et al. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in Quinolone-resistant Shigella dysenteriae serotype 1 clinical isolates from Kolkata, India. Antimicrob Agents Chemother 2005;49:1660-1. [Crossref] [PubMed]
- Chau TT, Campbell JI, Galindo CM, et al. Antimicrobial drug resistance of Salmonella enterica serovar typhi in asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother 2007;51:4315-23. [Crossref] [PubMed]
- Cavaco LM, Hasman H, Xia S, et al. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother 2009;53:603-8. [Crossref] [PubMed]
- Wang M, Guo Q, Xu X, et al. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother 2009;53:1892-7. [Crossref] [PubMed]
- Pu XY, Pan JC, Wang HQ, et al. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother 2009;63:917-20. [Crossref] [PubMed]
- Yamane K, Wachino J, Suzuki S, et al. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob Agents Chemother 2008;52:1564-6. [Crossref] [PubMed]
- Giedraitienė A, Vitkauskienė A, Naginienė R, et al. Antibiotic resistance mechanisms of clinically important bacteria. Medicina 2011;47:137-46. [Crossref] [PubMed]
- Sorlozano A, Gutierrez J, Jimenez A, et al. Contribution of a new mutation in parE to quinolone resistance in extended-spectrum-beta-lactamase-producing Escherichia coli isolates. J Clin Microbiol 2007;45:2740-2. [Crossref] [PubMed]
- Jennison AV, Verma NK. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev 2004;28:43-58. [Crossref] [PubMed]
- Livio S, Strockbine NA, Panchalingam S, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis 2014;59:933-41. [Crossref] [PubMed]
- Noriega FR, Liao FM, Maneval DR, et al. Strategy for cross-protection among Shigella flexneri serotypes. Infect Immun 1999;67:782-8. [PubMed]
- Xia S, Xu B, Huang L, et al. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J Clin Microbiol 2011;49:232-42. [Crossref] [PubMed]
- Pu XY, Pan JC, Zhang W, et al. Quinolone resistance-determining region mutations and the plasmid-mediated quinolone resistance gene qnrS played important roles in decreased susceptibility to fluoroquinolones among Shigella isolates in southeast China between 1998 and 2013. Int J Antimicrob Agents 2015;45:438-9. [Crossref] [PubMed]
- Yang H, Chen G, Zhu Y, et al. Surveillance of antimicrobial susceptibility patterns among Shigella species isolated in China during the 7-year period of 2005-2011. Ann Lab Med 2013;33:111-5. [Crossref] [PubMed]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell 2007;128:1037-50. [Crossref] [PubMed]
- Tamanna Ramana J. Structural Insights into the Fluoroquinolone Resistance Mechanism of Shigella flexneri DNA Gyrase and Topoisomerase IV. Microb Drug Resist 2016;22:404-11. [Crossref] [PubMed]
- Azmi IJ, Khajanchi BK, Akter F, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS One 2014;9. [Crossref] [PubMed]
- Yang C, Li P, Zhang X, et al. Molecular characterization and analysis of high-level multidrug-resistance of Shigella flexneri serotype 4s strains from China. Sci Rep 2016;6:29124. [Crossref] [PubMed]
- Cui X, Wang J, Yang C, et al. Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front Microbiol 2015;6:435. [Crossref] [PubMed]
- Vila J, Ruiz J, Marco F, et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother 1994;38:2477-9. [Crossref] [PubMed]
- Jacoby G, Cattoir V, Hooper D, et al. qnr Gene nomenclature. Antimicrob Agents Chemother 2008;52:2297-9. [Crossref] [PubMed]
- Poirel L, Cattoir V, Nordmann P. Plasmid-Mediated Quinolone Resistance; Interactions between Human, Animal, and Environmental Ecologies. Front Microbiol 2012;3:24. [Crossref] [PubMed]
- Luo Y, Yang J, Zhang Y, et al. Prevalence of β-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents 2011;37:352-5. [Crossref] [PubMed]