Alternative Splicing Detection Tool—a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis
Introduction
Next-generation sequencing (NGS) is expected to have a tremendous impact on genomic research. NGS methodology has the ability to sequence DNA or RNA fragments with extraordinary speed that has never been achieved before and therefore enables the development of original applications, accelerating novel scientific discoveries (1). In fact, genome sequencing projects that previously required numerous Sanger sequencing experiments can now be accomplished in a couple of days with a cost reduction of many thousand times, due to the massive parallel sequencing that NGS offers (2). NGS can be an extremely useful tool, used to uncover the complexity of the human genome as well as genetic alterations in a cancer genome, or specific single nucleotide polymorphisms (SNPs) that may possess a vital role in many diseases such as cancer. Since many genetic variants which contribute to many human conditions are still unknown, NGS will definitely contribute to the identification of these genetic variants, including single nucleotide variants (SNVs) or SNPs, small insertions and deletions (indels, 1–1,000 bp), structural and genomic variants (>1,000 bp) (3).
Except from genetic variations, NGS can also unveil information regarding alternative splicing events or transcript identifications (4,5). It should be noticed that with the aid of high-throughput sequencing, the identification of low-frequency variants is now feasible. This innovative ability that NGS technology provides is of high significance because the existence, or not, of a specific splice variant constitutes vital information with potential clinical value in many diseases. For example, studies in LnCaP cell line revealed that the overexpression of short TRMP8α (sM8α), a specific alternative transcript of transient receptor potential melastatin 8 (TRPM8), not only can lead to the promotion of cell migration and invasion, but also to the mitigation of starvation-induced apoptosis (6). In addition, high-throughput sequencing technology of NGS can divulge various alternative transcripts with predictive value which could be used as potential biomarkers, thus enabling a better diagnosis or prognosis for many diseases. Recent evidence regarding the B cell CLL/lymphoma 11A (BCL11A) gene, revealed that a specific transcript, BCL11A-XL, is an independent predictor of relapse in cases of squamous cell carcinomas and large cell carcinomas, thus possessing biomarker capabilities (7).
Despite its tremendous power and significant advantages, NGS demonstrates two serious limitations: short reads that provide information for a limited nucleotide region as well as the massive output data that is produced. Each NGS run can provide researchers with an enormous amount of raw data that makes both storage and analysis hard to accomplish. These facts suggest that advanced bioinformatics tools have become not only a necessity, but a priority, in order for NGS technology to be used efficiently and solve unanswered questions on genomic research that previous technologies were incapable of solving (8). The manipulation and analysis of sequencing data acquired by NGS platforms regarding alternative splicing events are probably the most challenging part of the whole sequencing workflow. For this reason, we designed a suitable algorithm that efficiently and automatically detects and identifies alternative splicing events that exist in high-throughput sequencing data. As a result, our algorithm “Alternative Splicing Detection Tool (ASDT)” is a script written in PERL that is capable of identifying alternative splicing events, providing the user with both qualitative and quantitative results regarding alternative splicing in the obtained NGS data. ASDT is available as a PERL script at http://aias.biol.uoa.gr/~mtheo, along with ReadMe.pdf file that contains detailed instructions on how to run ASDT as well as example input files for a tutorial run.
Necessary input files for ASDT
ASDT was designed to detect both annotated and novel alternative splicing events in high-throughput NGS data along with their frequency, providing the number of sequencing reads corresponding to each splicing event. In addition, ASDT requires two input files for a successful run. The first input file is the full nucleotide GenBank® report of the gene of interest that can be downloaded from the GenBank® sequence database, which is an open-access, annotated collection of all publicly available nucleotide sequences and the respective proteins. This file contains information about all the annotated messenger RNAs (mRNAs), miscellaneous RNAs (miscRNAs) as well as non-coding RNAs (ncRNAs) that are transcribed from the gene of interest. Detailed instructions on how GenBank® report file can be downloaded are shown in ReadMe.pdf. The second input file is the raw data of the NGS run (FASTQ file). FASTQ format is a text-based format for storing both a nucleotide sequence and its corresponding quality scores. In FASTQ files each record consists of four consecutive lines. The first line is the ID of the sequencing read, the second line is the sequence read, the third line contains the symbol “+” and therefore does not have any biological and experimental significance, whereas the fourth line indicates the quality score for each nucleotide of the second line.
Analysis of alternative splicing events
Each ASDT run can be separated in two major parts. The first part of the analysis corresponds to the detection of annotated and novel alternative splicing events, providing the number of sequencing reads that characterize them. During this part of the analysis, ASDT uses the first input file to extract all the exon coordinates, generating in this way all exon and intron sequences from all annotated transcripts (mRNAs, miscRNAs or ncRNAs) of the gene. Then, it synthesizes nucleotide keywords for every possible alternative splicing event between all annotated exons, merging a number of bases from the 3’-end (donor) of each exon with a number of bases from the 5’-start (acceptor) of all exons that follow downstream. For each keyword that is produced, a reverse complement keyword is also synthesized, so that results regarding both strands are collected. When all nucleotide keywords are formed, ASDT tests the existence of each keyword—which in fact represents a specific splice junction—in the FASTQ file. If a match is detected, ASDT extracts in a separate file (All reads.txt) the sequence identifier, the sequencing read that represents it as it appears in the FASTQ file, the keyword that was matched and the exons that formed it (Figure 1A). Apart from reads with a simple match—therefore containing only one splicing event—ASDT also extracts in a separate output file (Multiple alternative splicing events.txt) all the sequencing reads that match multiple nucleotide keywords, thus containing more than one splice junctions (Figure 1B).
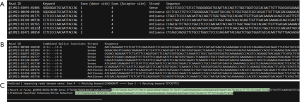
Detection of novel cryptic exons, exon extensions or intron retentions
Regarding the second (optional) part of the analysis, ASDT can also analyze the intronic regions of the gene(s), thus enabling the detection of novel cryptic exons residing in annotated introns, extensions of previously annotated exons, or even intron retentions. During this part of the analysis, ASDT splits every intron sequence with a specific step of nucleotides, which is provided by the user in order to synthesize intronic nucleotide keywords that will cover the entire intron. Moreover, alongside every newly formed intronic sequence a reverse complement keyword will be also produced, similarly to the first part. Then, every nucleotide keyword is tested whether or not it appears in the sequencing reads of the obtained FASTQ file. Finally, each matching read that is predicted to contain novel information regarding a potential novel cryptic exon, exon extension or intron retention is printed in a separate output file, Prediction of exons-Exon extensions-Intron Retentions.txt (Figure 1C).
Parameter modifications using ASDT
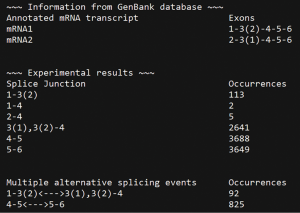
ASDT provides a user-friendly environment during each run, enabling the user to modify from the command prompt many searching parameters of the run, in order to get more accurate and personalized results. For instance, the number of nucleotides that will form each keyword is chosen by the user through the command prompt. Additionally, it gives the opportunity to the user to search each nucleotide keyword in the FASTQ file not only with perfect matches, but also with mismatches, enabling the identification of splice junctions even in sequencing reads with low sequencing quality containing mismatched base(s) in the splice junction regions. The searching option using mismatches as well as the number of mismatches that will be applied in the analysis is also decided by the user through the command prompt. Furthermore, all alternative splicing events that were detected are summarized in a separate output file, providing a simple and quick overview of all the alternative splice junctions that were detected, including novel ones, as well as the number of occurrences each one was noticed (Figure 2).
Comparison of ASDT with other similar algorithms or software
Despite its power, NGS has made the research of alternative splicing hard to accomplish, due to the massive amount of data and the short sequencing reads it produces. Hundreds, thousands or even millions of short reads can be generated in experimental runs of every NGS platform (1). This fact makes the detection of rare alternative splicing events an extremely challenging procedure. As it can be concluded by many recent studies regarding alternative splicing, many novel splice junctions are significantly rare in number, compared with the annotated ones (9). In this way, their detection in NGS data without a specific tool or software is not feasible. For this reason, there have been many computational approaches and algorithms designed specifically for the analysis of alternative splicing from NGS data, but they demonstrate major differences with ASDT.
A recent software tool, Splicing Express, is able to identify patterns of simple alternative splicing events, such as exon skipping, intron retention and alternative 5' and 3'splicing borders, but it requires GTF files resulting from transcriptome assembly in order to manipulate the NGS data (10). As a result, users have to run Tophat2 or another read-aligner program and then a transcriptome assembly tool, before running Splicing Express. Other widely used bioinformatics tools for alternative splicing analysis include ABMapper (11), Comrad (12) and GSNAP (13). However, these tools demonstrate major differences with ASDT as well. ABMapper is specifically designed for exploring all putative locations of reads that are mapped to splice junctions, by using two seeds from each end of a read and searches for canonical splicing motifs (i.e., ‘GT-AG’, ‘GC-AG’ and ‘AT-AC’) to determine a stop point for spliced alignment. In addition, Comrad is designed to analyze RNA-Seq and whole genome shotgun sequencing (WGSS) data for the purposes of discovering genomic rearrangements and aberrant transcripts and therefore provides accurate classification of rearrangements as expressed or not expressed and accurate classification of the genomic or non-genomic origin of aberrant transcripts. Finally, GSNAP can detect short and long-distance splicing, including interchromosomal splicing, in individual reads, but unlike ASDT, it uses probabilistic models or a database of known splice sites. However, to the best of our knowledge ASDT is the first tool that is designed for alternative splicing analysis using only FASTQ files. Therefore, we believe that ASDT possesses some innovative and unique features that are not available in any other publicly accessible resource and has the capacity to efficiently contribute to the alternative splicing analysis of NGS data. ASDT analyzes alternative splicing events without the need for online software tools or even spliced read mappers for RNA-Seq, like TopHat2 (14). Another important feature of ASDT is its ability to detect low-frequency splice junctions, even with unique representation in the FASTQ file. This property renders this tool more sensitive than other algorithms that are currently used for this purpose. In addition, ASDT enables alternative splicing analysis without the need for comparing the annotation from GTF/GFF file, in order to identify the novel alternative splicing sites. Furthermore, users do not have to generate additional BAM/BED files and proceed to visualization using genome browsers, like UGENE (15) and IGV (16). In addition to the aforementioned, except from alternative splicing events, ASDT gives the opportunity for the user to detect potential novel exons, exon extensions or even intron retentions for the gene of interest, providing in our opinion a quick, efficient and comprehensive view of the experimental results derived from the NGS data.
Biological and clinical significance of splice variants
To date, a lot of genes are known to express alternative splice variants, some of which may have significant clinical value (17-27). The family of KLKs constitutes such a prominent example (28,29), as particular splice variants of the KLK1–KLK15 genes have been highlighted as putative biomarkers in human malignancies (30-35). Very recently, ASDT has been used to partially reveal novel transcripts of tissue kallikrein (KLK1) (36) and several other members of the kallikrein-related peptidase (KLK) family (36-39), as well as splice variants of the BCL2L12 gene (40,41), a prominent member of the apoptosis-related BCL2 family (42,43) with important prognostic value both in regard with solid tumors (44-46) as well as hematological malignancies (47,48), similarly to the major members of the BCL2 family, namely BCL2 and BAX (49-51). Moreover, the expression of a splice variant could be combined with clinical markers; for instance, the expression of BCL2L12 splice variant 1 and BCL2 variant alpha in malignant B cells of patients with chronic lymphocytic leukemia could be combined with lymphocyte count and/or other clinical features to provide a better prognostic system than the currently used ones (48,52). Similarly, expression of important transcripts resulting from alternative splicing in blasts of patients with myelodysplastic syndromes could be used along with novel markers such as monosomal karyotype to predict prognosis (53), treatment decision-making (54), or patients’ response to therapy (55,56), at is has recently been proposed.
Distinct 3’-untranslated regions (3’-UTRs) of splice variants of a single protein-coding gene resulting from alternative splicing may have other implications, including different regulatory properties. As microRNAs (miRNAs) bind 3’-UTRs to downregulate the levels of the respective proteins or protein isoforms, alterations in their sequence due to alternative splicing may account for the regulation of splice variants by distinct miRNAs (57). Thus, miRNAs regulating the expression of protein-coding transcripts subjected to intense alternative splicing may constitute very promising molecular biomarkers; such miRNAs include members of the miR-17/92 cluster (58) and its paralogue miR-106a/363, such as miR-92a-3p and miR-20b-5p as well as miR-155-5p, which have pivotal roles in other B-cell malignancies, including chronic lymphocytic leukemia (59-61). Other such important biomarkers regulating splice variants of protein-coding genes and being clinically significant include miR-15a-5p, miR-16 miR-34a, and miR-96 regulating BCL2 expression (62-65), miR-182 regulating BCL2L12 expression (66), miR-200 family members (67), as well as several other miRNAs (68-71).
Conclusions
As it can be easily assumed, given the vast amount of data produced by NGS, developing a massive data storage and management solution and creating bioinformatic tools to effectively analyze it will be the most crucial step for the successful application of NGS methodology (72). The full benefit of NGS will not be achieved until bioinformatics are able to maximally utilize and interpret these enormous short-read sequences into molecular and genetic useful information (73). Consequently, we believe that ASDT demonstrates many innovative and unique characteristics that can efficiently analyze alternative splicing events or identify novel cryptic exons, exon extensions or intron retentions from high-throughput NGS data.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Zhang J, Chiodini R, Badr A, et al. The impact of next-generation sequencing on genomics. J Genet Genomics 2011;38:95-109. [Crossref] [PubMed]
- Radovich M. Next-generation sequencing in breast cancer: translational science and clinical integration. Pharmacogenomics 2012;13:637-9. [Crossref] [PubMed]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 2001;17:100-7. [Crossref] [PubMed]
- Bryant DW Jr, Priest HD, Mockler TC. Detection and quantification of alternative splicing variants using RNA-seq. Methods Mol Biol 2012;883:97-110. [Crossref] [PubMed]
- Park JW, Tokheim C, Shen S, et al. Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol Biol 2013;1038:171-9. [Crossref] [PubMed]
- Peng M, Wang Z, Yang Z, et al. Overexpression of short TRPM8 variant alpha promotes cell migration and invasion, and decreases starvation-induced apoptosis in prostate cancer LNCaP cells. Oncol Lett 2015;10:1378-84. [Crossref] [PubMed]
- Zhang N, Jiang BY, Zhang XC, et al. The BCL11A-XL expression predicts relapse in squamous cell carcinoma and large cell carcinoma. J Thorac Dis 2015;7:1630-6. [PubMed]
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 2000;103:367-70. [Crossref] [PubMed]
- Adamopoulos PG, Raptis GD, Kontos CK, et al. Discovery and expression analysis of novel transcripts of the human SR-related CTD-associated factor 1 (SCAF1) gene in human cancer cells using Next-Generation Sequencing. Gene 2018;670:155-65. [Crossref] [PubMed]
- Kroll JE, Kim J, Ohno-Machado L, et al. Splicing Express: a software suite for alternative splicing analysis using next-generation sequencing data. PeerJ 2015;3. [Crossref] [PubMed]
- Lou SK, Ni B, Lo LY, et al. ABMapper: a suffix array-based tool for multi-location searching and splice-junction mapping. Bioinformatics 2011;27:421-2. [Crossref] [PubMed]
- McPherson A, Wu C, Hajirasouliha I, et al. Comrad: detection of expressed rearrangements by integrated analysis of RNA-Seq and low coverage genome sequence data. Bioinformatics 2011;27:1481-8. [Crossref] [PubMed]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873-81. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Okonechnikov K, Golosova O, Fursov M, et al. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 2012;28:1166-7. [Crossref] [PubMed]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24-6. [Crossref] [PubMed]
- Kontos CK, Adamopoulos PG, Scorilas A. Molecular biomarkers of laryngeal cancer. In: Biomarkers in disease: Methods, discoveries and applications: Biomarkers in cancer. Netherlands: Spring, 2015:891-919.
- Kontos CK, Adamopoulos PG, Scorilas A. Prognostic and predictive biomarkers in prostate cancer. Expert Rev Mol Diagn 2015;15:1567-76. [Crossref] [PubMed]
- Kontos CK, Scorilas A, Papavassiliou AG. The role of transcription factors in laboratory medicine. Clin Chem Lab Med 2013;51:1563-71. [PubMed]
- Geomela PA, Kontos CK, Yiotakis I, et al. L-DOPA decarboxylase mRNA expression is associated with tumor stage and size in head and neck squamous cell carcinoma: a retrospective cohort study. BMC Cancer 2012;12:484. [Crossref] [PubMed]
- Kontos CK, Papadopoulos IN, Fragoulis EG, et al. Quantitative expression analysis and prognostic significance of L-DOPA decarboxylase in colorectal adenocarcinoma. Br J Cancer 2010;102:1384-90. [Crossref] [PubMed]
- Kontos CK, Papageorgiou SG, Diamantopoulos MA, et al. mRNA overexpression of the hypoxia inducible factor 1 alpha subunit gene (HIF1A): An independent predictor of poor overall survival in chronic lymphocytic leukemia. Leuk Res 2017;53:65-73. [Crossref] [PubMed]
- Psyrri A, Papageorgiou S, Liakata E, et al. Phosphatidylinositol 3'-kinase catalytic subunit alpha gene amplification contributes to the pathogenesis of mantle cell lymphoma. Clin Cancer Res 2009;15:5724-32. [Crossref] [PubMed]
- Vassilakopoulou M, Avgeris M, Velcheti V, et al. Evaluation of PD-L1 Expression and Associated Tumor-Infiltrating Lymphocytes in Laryngeal Squamous Cell Carcinoma. Clin Cancer Res 2016;22:704-13. [Crossref] [PubMed]
- Kefala M, Papageorgiou SG, Kontos CK, et al. Increased expression of phosphorylated NBS1, a key molecule of the DNA damage response machinery, is an adverse prognostic factor in patients with de novo myelodysplastic syndromes. Leuk Res 2013;37:1576-82. [Crossref] [PubMed]
- Miltiades P, Lamprianidou E, Vassilakopoulos TP, et al. The Stat3/5 Signaling Biosignature in Hematopoietic Stem/Progenitor Cells Predicts Response and Outcome in Myelodysplastic Syndrome Patients Treated with Azacitidine. Clin Cancer Res 2016;22:1958-68. [Crossref] [PubMed]
- Kontos CK. Surrogate Prognostic Biomarkers in OSCC: The Paradigm of PA28gamma Overexpression. EBioMedicine 2015;2:784-5. [Crossref] [PubMed]
- Kontos CK, Mavridis K, Talieri M, et al. Kallikrein-related peptidases (KLKs) in gastrointestinal cancer: mechanistic and clinical aspects. Thromb Haemost 2013;110:450-7. [Crossref] [PubMed]
- Kontos CK, Scorilas A. Kallikrein-related peptidases (KLKs): a gene family of novel cancer biomarkers. Clin Chem Lab Med 2012;50:1877-91. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Papageorgiou SG, et al. KLKB1 mRNA overexpression: A novel molecular biomarker for the diagnosis of chronic lymphocytic leukemia. Clin Biochem 2015;48:849-54. [Crossref] [PubMed]
- Alexopoulou DK, Kontos CK, Christodoulou S, et al. KLK11 mRNA expression predicts poor disease-free and overall survival in colorectal adenocarcinoma patients. Biomark Med 2014;8:671-85. [Crossref] [PubMed]
- Christodoulou S, Alexopoulou DK, Kontos CK, et al. Kallikrein-related peptidase-6 (KLK6) mRNA expression is an independent prognostic tissue biomarker of poor disease-free and overall survival in colorectal adenocarcinoma. Tumour Biol 2014;35:4673-85. [Crossref] [PubMed]
- Foteinou E, Kontos CK, Giotakis AI, et al. Low mRNA expression levels of kallikrein-related peptidase 4 (KLK4) predict short-term relapse in patients with laryngeal squamous cell carcinoma. Biol Chem 2014;395:1051-62. [Crossref] [PubMed]
- Kontos CK, Adamopoulos PG, Papageorgiou SG, et al. mRNA overexpression of kallikrein-related peptidase 14 (KLK14) is an independent predictor of poor overall survival in chronic lymphocytic leukemia patients. Clin Chem Lab Med 2016;54:315-24. [Crossref] [PubMed]
- Kontos CK, Chantzis D, Papadopoulos IN, et al. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett 2013;330:106-12. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Discovery of novel transcripts of the human tissue kallikrein (KLK1) and kallikrein-related peptidase 2 (KLK2) in human cancer cells, exploiting Next-Generation Sequencing technology. Genomics 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Molecular cloning of novel transcripts of human kallikrein-related peptidases 5, 6, 7, 8 and 9 (KLK5 - KLK9), using Next-generation sequencing. Sci Rep 2017;7:17299. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Identification and molecular cloning of novel transcripts of the human kallikrein-related peptidase 10 (KLK10) gene using next-generation sequencing. Biochem Biophys Res Commun 2017;487:776-81. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Novel splice variants of the human kallikrein-related peptidases 11 (KLK11) and 12 (KLK12), unraveled by next-generation sequencing technology. Biol Chem 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Tsiakanikas P, et al. Identification of novel alternative splice variants of the BCL2L12 gene in human cancer cells using next-generation sequencing methodology. Cancer Lett 2016;373:119-29. [Crossref] [PubMed]
- Kontos CK, Scorilas A. Molecular cloning of novel alternatively spliced variants of BCL2L12, a new member of the BCL2 gene family, and their expression analysis in cancer cells. Gene 2012;505:153-66. [Crossref] [PubMed]
- Christodoulou MI, Kontos CK, Halabalaki M, et al. Nature promises new anticancer agents: Interplay with the apoptosis-related BCL2 gene family. Anticancer Agents Med Chem 2014;14:375-99. [Crossref] [PubMed]
- Kontos CK, Christodoulou MI, Scorilas A. Apoptosis-related BCL2-family members: Key players in chemotherapy. Anticancer Agents Med Chem 2014;14:353-74. [Crossref] [PubMed]
- Fendri A, Kontos CK, Khabir A, et al. BCL2L12 is a novel biomarker for the prediction of short-term relapse in nasopharyngeal carcinoma. Mol Med 2011;17:163-71. [Crossref] [PubMed]
- Geomela PA, Kontos CK, Yiotakis I, et al. Quantitative expression analysis of the apoptosis-related gene, BCL2L12, in head and neck squamous cell carcinoma. J Oral Pathol Med 2013;42:154-61. [Crossref] [PubMed]
- Kontos CK, Papadopoulos IN, Scorilas A. Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene BCL2L12 in colon cancer. Biol Chem 2008;389:1467-75. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Foukas PG, et al. BCL2L12 protein overexpression is associated with favorable outcome in diffuse large B-cell lymphoma patients in the rituximab era. Leuk Lymphoma 2016;57:2199-203. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Pappa V, et al. The novel member of the BCL2 gene family, BCL2L12, is substantially elevated in chronic lymphocytic leukemia patients, supporting its value as a significant biomarker. Oncologist 2011;16:1280-91. [Crossref] [PubMed]
- Fendri A, Kontos CK, Khabir A, et al. Quantitative analysis of BCL2 mRNA expression in nasopharyngeal carcinoma: an unfavorable and independent prognostic factor. Tumour Biol 2010;31:391-9. [Crossref] [PubMed]
- Giotakis AI, Kontos CK, Manolopoulos LD, et al. High BAX/BCL2 mRNA ratio predicts favorable prognosis in laryngeal squamous cell carcinoma, particularly in patients with negative lymph nodes at the time of diagnosis. Clin Biochem 2016;49:890-6. [Crossref] [PubMed]
- Kontos CK, Fendri A, Khabir A, et al. Quantitative expression analysis and prognostic significance of the BCL2-associated X gene in nasopharyngeal carcinoma: a retrospective cohort study. BMC Cancer 2013;13:293. [Crossref] [PubMed]
- Mpakou VE, Ioannidou HD, Konsta E, et al. Quantitative and qualitative analysis of regulatory T cells in B cell chronic lymphocytic leukemia. Leuk Res 2017;60:74-81. [Crossref] [PubMed]
- Papageorgiou SG, Vasilatou D, Kontos CK, et al. The prognostic value of monosomal karyotype (MK) in higher-risk patients with myelodysplastic syndromes treated with 5-Azacitidine: A retrospective analysis of the Hellenic (Greek) Myelodysplastic syndromes Study Group. Am J Hematol 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Sterlacci W, Sioulas AD, Veits L, et al. 22-gauge core vs 22-gauge aspiration needle for endoscopic ultrasound-guided sampling of abdominal masses. World J Gastroenterol 2016;22:8820-30. [Crossref] [PubMed]
- Papageorgiou SG, Vasilatou D, Kontos CK, et al. Treatment with 5-Azacytidine improves clinical outcome in high-risk MDS patients in the 'real life' setting: A single center observational study. Hematology 2016;21:34-41. [Crossref] [PubMed]
- Tarushi A, Raptopoulou CP, Psycharis V, et al. Copper(II) Inverse-[9-Metallacrown-3] Compounds Accommodating Nitrato or Diclofenac Ligands: Structure, Magnetism, and Biological Activity. Eur J Inorg Chem 2016;2016:219-31. [Crossref]
- Kontos CK, Vasilatou D, Papageorgiou SG, et al. Translation Regulation by microRNAs in Acute Leukemia. In: Reviews in Cell Biology and Molecular Medicine. Wiley-VCH Verlag GmbH & Co. KgaA, 2014.
- Vasilatou D, Papageorgiou SG, Kontsioti F, et al. Expression analysis of mir-17-5p, mir-20a and let-7a microRNAs and their target proteins in CD34+ bone marrow cells of patients with myelodysplastic syndromes. Leuk Res 2013;37:251-8. [Crossref] [PubMed]
- Papageorgiou SG, Diamantopoulos MA, Kontos CK, et al. MicroRNA-92a-3p overexpression in peripheral blood mononuclear cells is an independent predictor of prolonged overall survival of patients with chronic lymphocytic leukemia. Leuk Lymphoma 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Tsiakanikas P, et al. Elevated miR-20b-5p expression in peripheral blood mononuclear cells: A novel, independent molecular biomarker of favorable prognosis in chronic lymphocytic leukemia. Leuk Res 2018;70:1-7. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Diamantopoulos MA, et al. MicroRNA-155-5p Overexpression in Peripheral Blood Mononuclear Cells of Chronic Lymphocytic Leukemia Patients Is a Novel, Independent Molecular Biomarker of Poor Prognosis. Dis Markers 2017;2017. [Crossref] [PubMed]
- Diamantopoulos MA, Kontos CK, Kerimis D, et al. Upregulated miR-16 expression is an independent indicator of relapse and poor overall survival of colorectal adenocarcinoma patients. Clin Chem Lab Med 2017;55:737-47. [Crossref] [PubMed]
- Kontos CK, Tsiakanikas P, Avgeris M, et al. miR-15a-5p, A Novel Prognostic Biomarker, Predicting Recurrent Colorectal Adenocarcinoma. Mol Diagn Ther 2017;21:453-64. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Christodoulou S, et al. miR-34a overexpression predicts poor prognostic outcome in colorectal adenocarcinoma, independently of clinicopathological factors with established prognostic value. Clin Biochem 2017;50:918-24. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Papadopoulos IN, et al. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumour Biol 2016;37:11815-24. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Papadopoulos IN, et al. Enhanced miR-182 transcription is a predictor of poor overall survival in colorectal adenocarcinoma patients. Clin Chem Lab Med 2014;52:1217-27. [Crossref] [PubMed]
- Skourti E, Logotheti S, Kontos CK, et al. Progression of mouse skin carcinogenesis is associated with the orchestrated deregulation of mir-200 family members, mir-205 and their common targets. Mol Carcinog 2016;55:1229-42. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Rapti SM, et al. miR-224 overexpression is a strong and independent prognosticator of short-term relapse and poor overall survival in colorectal adenocarcinoma. Int J Oncol 2015;46:849-59. [Crossref] [PubMed]
- Ferraro A, Kontos CK, Boni T, et al. Epigenetic regulation of miR-21 in colorectal cancer: ITGB4 as a novel miR-21 target and a three-gene network (miR-21-ITGBeta4-PDCD4) as predictor of metastatic tumor potential. Epigenetics 2014;9:129-41. [Crossref] [PubMed]
- Kerimis D, Kontos CK, Christodoulou S, et al. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin Biochem 2017;50:285-92. [Crossref] [PubMed]
- Tsiakanikas P, Kontos CK, Kerimis D, et al. High microRNA-28-5p expression in colorectal adenocarcinoma predicts short-term relapse of node-negative patients and poor overall survival of patients with non-metastatic disease. Clin Chem Lab Med 2018;56:990-1000. [Crossref] [PubMed]
- Parmakelis A, Kotsakiozi P, Kontos CK, et al. The transcriptome of a "sleeping" invader: de novo assembly and annotation of the transcriptome of aestivating Cornu aspersum. BMC Genomics 2017;18:491. [Crossref] [PubMed]
- Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet 2008;24:142-9. [Crossref] [PubMed]


