Precision oncology in liver cancer
Introduction
Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) are the two most common forms of primary liver cancer (PLC); together, they are the third most common cause of cancer-related death worldwide. According to the Global Burden of Disease Study, in 2015 there were over 850,000 incident cases of liver cancer globally, and over 800,000 deaths (1). While the age-standardized mortality attributable to liver cancer has significantly decreased over the past several decades in many high-prevalence regions including Asia, Africa and South America, Western countries such as the United States and Canada have experienced increases. Chronic liver disease predisposes patients to develop HCC; almost 85% of cases result from arguably preventable causes such as alcohol and viral hepatitis. We thus expect that with improvements in health education, practice, policy and prevention, the global burden of liver cancer driven by these risk factors may continue to reduce. For example, anti-viral treatments have become highly effective and widely available. Direct antiviral agents (DAA) against HCC have made it possible for the majority of chronically-infected patients to clear the virus, with subsequent stabilization or improvement in liver health (2,3). Unfortunately, some studies have reported that the eradication of HCV by DAA may promote new or recurrent HCC (4-7). While this finding has not been borne out in larger studies, the debate continues regarding the early impact of DAA on HCC (8-11). Nonetheless, it is prudent to continue to monitor and surveil HCV-treated livers for the development of PLC until longer term studies can provide better evidence to establish guidelines for more accurate and efficient screening of the HCV population.
In contrast to the decreasing incidence of HCV-induced HCCs, tumors driven by non-alcoholic fatty liver disease (NAFLD), which is associated with the metabolic syndrome, are becoming more prevalent. As the obesity epidemic continues to climb worldwide, a much larger population will be at risk for developing liver cancer (12,13), thus new strategies are urgently needed to address this formidable epidemic (14).
Unlike most other solid tumors, PLCs respond poorly to conventional chemotherapy such that no agents are currently FDA-approved for the treatment of advanced HCC; rather several tyrosine kinase inhibitors (TKIs) have demonstrated efficacy in patients with preserved liver function (15). Both sorafenib and regorafenib are non-specific kinase inhibitors that are currently used in HCC, and their clinical benefits extend survival by a mere 2–3 months. The efficacy of these drugs suggests that aberrant kinase activities are relevant in HCC, but yet their poor response rates highlight a significant deficiency in our understanding of the role of specific kinases and other potential therapeutic targets in HCC.
Tumor heterogeneity & limitations of molecular profiling
Widespread application of next-generation sequencing (NGS) has led to detailed molecular characterization of HCC and ICC (16-21). These studies confirm that HCC and ICC are genetically distinct, with recurrent mutations in KRAS, IDH1/2, FGFR, EGFR-ERBB in ICC, as compared to Wnt-βcatenin, TERT, JAK-STAT, and PI3K-AKT-mTORC1 mutations in HCC. Despite these frequent mutations, each tumor contains many more somatic mutations (up to hundreds) that may contribute to its pathogenesis. Therefore, each cancer bears a unique genetic signature, which continues to evolve as the cancer progresses and gives rise to heterogeneity within that tumor. Such complexity poses a major roadblock given our current approach to precision oncology, which focuses on one target at a time.
‘Druggable’ targets in ICC include IDH1/2, FGFR2/4, EGFR/ERBB2 and PIK3CA, but studies of molecularly targeted agents in these patients had mixed results. A preliminary ‘basket’ trial evaluating compounds targeting FGFR, IDH1, MET and MEK in biliary tract cancers reported that only 4 out of 13 patients responded to treatment (22). In a sub-analysis of ICCs included in the MOSCATO-01 trial, 23 out of 34 patients undergoing molecular profiling were found to have a potentially ‘actionable’ mutation; among them, 18 received targeted therapies, with an overall response rate of 33% (23). Other clinical trials reported similarly inconclusive benefits based on molecular findings (24), supporting the notion that we are far from effective precision oncology for patients with ICC.
A large number of kinase inhibitors have been used in patients with advanced HCC; only a few agents have demonstrated efficacy: sorafenib, levatinib, and regorafenib. Interestingly all of these agents are relatively non-specific in terms of their kinase targets, and many of these targets are involved in angiogenesis (i.e., VEGFR1, VEGFR2, VEGFR3, PDGFR, RET). Therefore, anti-angiogenesis may represent an important means of HCC stabilization, but these agents may not be directly inducing cytotoxicity of the cancer cells. Currently, sorafenib remains first-line systemic treatment for advanced HCC, but only has an objective response rate of 2–3%. In a phase III non-inferiority trial, lenvatinib lead to a clinically meaningful improvement in outcome compared with sorafenib with a higher objective response rate of 24% (25); it is expected to be approved as an alternative first-line therapy in 2018. On the other hand, regorafenib, which is structurally similar to sorafenib, has been approved as a second-line option in patients who tolerated but progressed on sorafenib. In this subset of patients, regorafenib achieved an objective response in 11%, leading to a median overall survival of 10.6 months (26).
In contrast to the promiscuous TKIs discussed above, agents with more restricted substrate specificity, such as rapamycin analogs or FGFR and c-MET inhibitors, have not been shown to impact survival as first-line therapy for patients with advanced HCC. This lack of an effect is particularly intriguing given the frequency with which these pathways are affected in HCC based on molecular studies. Further, there are no specific predictors of response to any of the FDA-approved TKIs. The modest success of kinase inhibitors in HCC highlights the critical knowledge gap in our understanding of the molecular pathogenesis of HCC, and our inability to translate the knowledge that we do have to the bedside. Some specific challenges that impede the success of targeted therapies include (I) tumor heterogeneity, (II) a lack of predictive biomarkers, (III) the inability to distinguish ‘driver’ from ‘passenger’ mutations, (IV) involvement of multiple signaling pathways in carcinogenesis, and (V) the development of drug resistance. In this review, we discuss a number of approaches that address these hurdles, with the goal of improving our delivery of precision oncology to patients with PLC.
HCC is usually the end-product of chronic liver injury of various etiologies (viral, metabolic, alcohol) and comes in many different flavors of disease, each with a unique molecular profile and cancer biology (21). These genetic changes have been categorized into ‘baskets’ involving ~10 pathways with only a handful of these being currently ‘actionable’. In addition, extensive cross-talk exists between these pathways, such that upregulation of one pathway can compensate for drug-induced inhibition of another pathway to maintain cellular functions. It can be argued that our current state of knowledge does not account for the complex biology that takes place within each tumor. New methods are needed to understand the sum total of the underlying molecular alterations found in individual cancer. Precision medicine holds great promise as we strive to treat disease based on genetic variation, but to do so we must bridge the gaps between genetic information, biology, and clinical medicine.
General approaches to precision oncology
Since cancer is the result of genetic and epigenetic mishaps, it is reasonable to suggest that the first step in achieving success in precision oncology is to define the genomic alterations of individual tumors. In fact, obtaining this type of data is now common and widespread in clinical practice. From the provider’s perspective, however, molecular driven therapies have often failed to achieve the desired clinical outcome. We now know that targeting the same mutation in different tumors does not lead to consistent response. This phenomenon was demonstrated in a study using vemurafenib to treat a variety of non-melanoma cancers that shared the same mutation (BRAF V600E), and the response rates varied greatly between tumor types, arguing that this BRAF mutation has varying functional significance in different cancers (27). Understanding the relationship between specific oncogenic mutations, tumor histology, and patient history and outcome is of paramount importance in predicting and designing tailored therapies with the aim of inducing clinically-meaningful responses.
Successes of ‘targeted’ therapy for solid cancers to date include imatinib for gastrointestinal stromal tumors, trastuzumab for HER2-positive breast and gastric cancers, and EGFR inhibitors for subset of NSCLCs, all of which have been validated in large phase III randomized studies. Outside of the context of clinical trials, however, precision oncology has yet to produce the anticipated widespread improvements in patient outcome. To date, few randomized trials have compared molecularly-informed treatments with standard selection by providers. The SHIVA trial was designed to match a panel of well-known cancer mutations with 10 FDA-approved drugs (28). Eligible patients (n=293) with ‘actionable’ mutations were randomly assigned to molecularly-targeted drugs or treatment at their physician’s choice. At a median follow-up of 11.3 months, the median progression-free survival was 2.3 months for the experimental (molecular-targeted) group and 2.0 months for the control group (P=0.41). While this trail has many limitations, it highlights the need for additional well-designed, prospective studies that consider both histology and genomic profile. Examples of ‘basket’ trials that are ongoing include the NCI-sponsored MATCH (NCT02465060) and MPACT (NCT01827384) studies, the ASCO-sponsored TAPUR (NCT02693535) registration study, and a number of European and Asian studies. These trials should provide much needed insight into the relationship between tumor response and molecular profile. In PLCs, however, inhibition of the ‘actionable’ pathways that are commonly mutated (PI3K-AKT-mTOR, FGF, c-MET) have already been tested in clinical studies; none have demonstrated significant clinical benefit as first-line therapy (29). Moving beyond ‘basket’ trials, new strategies are needed to improve the outcome of patients with advanced HCC. Here, we discuss three potential approaches to address the barriers noted above.
Targeting a specific HCC subtype
Though no two cancers are exactly alike, experts agree that common driver mutations exist that are shared among tumors, allowing molecular subclassification. It follows that if one can fully delineate molecular pathogenesis of each subclass of HCC, we could design therapies for individual cancer type. One example of a distinct, yet homogenous subgroup of HCC is fibrolamellar HCC (FLC). FLC is unusual in that it occurs in children and young adults without a history of underlying liver disease. Genomic analyses have identified a common mutation in these rare cancers that involves an in-frame deletion of ~400 kb on chromosome 19, resulting in a novel fusion protein, DNAJ-PKAc (30). The mutant gene product replaces the first exon of the catalytic subunit of protein kinase A (PKAc) with the N-terminus of a heat shock protein, DNAJ (a.k.a. HSP40). No other recurrent genomic changes have been found in FLCs, and the overall mutation burden in these cancers is low, thus the modified PKAc appears to be the dominant oncogenic driver of these cancers (31-33). In fact, recent studies have shown that the expression of DNAJ-PKAc is necessary and sufficient to induce liver tumorigenesis in mice, but the mechanism behind this transformation is unknown (34,35).
Patients with FLC typically present late with bulky disease; up to 70% have lymph node involvement at the time of diagnosis, leading to a poor overall 5-year survival rate of ~35% in these otherwise healthy young people (36,37). The disease has a distinct histology defined by large polygonal cells with eosinophilic cytoplasm and prominent nucleoli that grow in clusters surrounded by lamella of fibrous stroma, hence the name fibrolamellar (38). Unlike ‘classic’ forms of HCC, FLC often metastasizes to lymph nodes, and can be associated with non-hepatic encephalopathy in its advanced stages. Further, they do not respond to conventional chemotherapies or targeted therapies such as sorafenib. Surgical resection is the only effective therapy for FLC, but is not curative once disease has spread outside the liver.
Intuitively, one would assume that a mutation involving a kinase that causes cancer would involve constitutive activation of its kinase activity to drive cellular transformation, as is the case for ABL, EGFR, BRAF, and SRC-mutated cancers. However, biochemical studies of DNAJ-PKAc show that the fusion kinase has similar basal kinase activity as the wild-type protein and remains under the regulation of its R subunits in a cAMP-dependent manner (39,40). In FLCs, the fusion kinase is overexpressed compared to wild-type PKAc due to higher basal promoter activity of DNAJB1, which leads to increased kinase activity in response to cAMP. In mice, over-expression of Dnajb1-Prkaca in the liver, but not a kinase-dead mutant, causes tumor formation (35). Little is presently known about the events downstream of DNAJ-PKAc, and there is no evidence that inhibition of global PKAc activity is safe or of therapeutic value. Due to the ubiquitous function of PKA, non-selective inhibition would likely lead to severe cellular and organ dysfunction, hence an alternative approach will be necessary for these patients. In preliminary studies using a novel cell model of FLC, we identified an important function of the heat-shock protein (HSP) scaffold in regulating growth-factor associated AKAP-PKAc signaling and chemoresistance (manuscript in preparation). We are currently exploring the utility of blocking HSP function to expose specific vulnerabilities of FLC cells to kinase inhibition. These and other studies that address the molecular biology and pharmacology of FLC will provide critical insights to finding effective therapies for this unique PLC.
Functional validation in genomic medicine
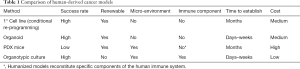
The majority of PLCs do not fall into genetically homogenous subtypes like FLC; rather they share many overlapping mutations across multiple pathways. It is impractical to study each tumor in extensive detail in order to understand the biologic nuances of their complex genomic disarray. An alternative strategy is to use a functional assay to test and validate targets that are identified through molecular profiling of individual cancers. Determining the relative efficacy of different compounds or combinations is particularly important when genomic profiling highlights more than 1 potential drug target, or when multiple drugs are available for a given target. Table 1 lists common methods used to query the biologic impact of genetic or pharmacologic manipulations in human cancers. While no one approach is perfect, each technique has advantages that can be exploited to investigate tumor biology and response to treatment. Recent advances allow propagation of human cancers either in vitro or in surrogate hosts. For example, ‘conditional reprogramming’, which entails co-culture of tumor cells with irradiated fibroblasts and a Rho kinase inhibitor greatly enhances the ability of primary human cells to be maintained in vitro (41). This technique has led to the development of many human cancer cell lines, as outlined in the Cancer Cell Line Encyclopedia. While these cell lines represent a valuable resource for large-scale screening, it is unclear whether these cells are representative of the diversity of cancer cells within one tumor following a period of in vitro selection. As such, resultant clones may not represent the heterogeneity that exists within the primary cancer (42).
Full table
3D-organoid cultures have recently attracted a great deal of attention given their ease of propagation and close resemblance of the physiological niche found in vivo. Refined protocols have improved the efficiency and speed of generating primary tumor spheroids, which can then be used for molecular analyses and drug screening. Broutier et al. describe a novel system of long-term in vitro propagation of PLCs that preserves their histologic architecture, gene expression, and genomic alterations (43). Using these organoids in drug screens, they identified ERK as a potential therapeutic target in HCC, but did not specify the specific patient subtype for which these agents could be effective (43). Pauli et al. reported the use of a similar approach to identify effective drugs for individual cancers (44); the predictive value of this method in clinical care remains to be proven in prospective studies.
A major drawback of 2D and 3D cultures is the absence of the native tumor microenvironment, which is known to play a critical role in tumor development and response to therapy. This deficit is particularly relevant in ICCs, in which a desmoplastic stromal reaction is often seen adjacent to tumor cells. In fact, the phenotype of cancer-associated fibroblasts has been associated with more advanced stage and poorer 5-year survival in these patients (45). Patient-derived xenografts (PDX) in immuno-deficient mice were created in an attempt to overcome this shortcoming of purely in vitro systems. Recent evidence suggests that the murine stroma adopts a metabolic phenotype that resembles the human cancer microenvironment (46), highlighting the importance of tumor-stromal interaction. While engraftment efficiency varies among tumor types, once established, PDX models recapitulate many features of primary human cancers including histology, gene expression, and drug response. A major limitation of the use of PDX models in clinical precision oncology is the long lag period necessary for engraftment and passaging, rendering this approach impractical for individuals seeking tailored cancer treatment. Secondary concerns about PDX models include selection bias in favor of more advanced tumors with high-grade features (47,48), variable engraftment efficiencies (49), and the high costs of PDX development, maintenance, and testing. In our experience, both HCCs and ICCs engraft subcutaneously in NSG mice, albeit at lower efficiency than colorectal cancers (unpublished data). At present, PLC PDX models are not readily available from commercial sources such as JAX or the Living Tumor Laboratory, therefore access is limited to a few investigators.
Organotypic slice cultures were originally described decades ago in neurophysiology studies (50,51), but have recently gained popularity in the study of human solid tumors. Tumor slice cultures (TSC) have multiple advantages, including methodologic simplicity and preservation of the tumor architecture. It has been used to assess relative drug sensitivity (52). Additional benefits of a standardized TSC platform are a high success rate, minimal lag period, and low cost. In our experience of >150 cases, viable TSCs have been generated from ~90% of tumors, and can be maintained in vitro for weeks, allowing for extensive experimental manipulation. Figure 1 demonstrates PLC TSCs that show variable responses to drug treatments. Since there is no in vitro selection, TSCs maintain tumor heterogeneity including the immune environment that is intrinsic to the human disease (53). Despite the advantages listed above, TSCs are limited by their finite lifespan without the ability for renewal.
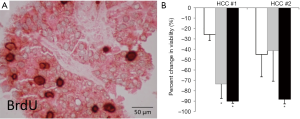
In summary, these ‘functional’ assays of human cancers fill an important gap in our ability to provide biologically relevant molecular data to guide therapy. We still struggle to make these assays readily available to patients, however, as most remain in the realm of research investigation, and none have been subjected to the rigor of a prospective clinical trial. Given these limitations, significant refinements in technique, data analyses and integration will be needed to allow us to realize the full potential of precision oncology.
Immunotherapies for HCC
Predictions by scientists over half a century ago suggested that a patient’s own immune system could selectively target and eliminate cancer cells from the body (54). This form of precision oncology has finally gained wide application with the use of immune checkpoint inhibitors in treating cancers such as melanoma, urothelial carcinoma, and NSCLC. Recent FDA-approval of the anti-programmed cell death protein 1 (PD1) monoclonal antibody, pembrolizumab, marks the first time a drug has been recommended based on a biomarker predictive of mutational burden irrespective of tumor histology (55). In addition to PLCs with microsatellite unstable phenotype, interest in immunotherapy for HCC has been spurred by cases of spontaneous regression in the setting of systemic inflammatory responses (56). Immune checkpoint inhibitors target molecules such as cytotoxic T-lymphocyte protein 4 (CTLA-4), PD-1, or programmed cell death ligand 1 (PD-L1). The end effect of antibody-mediated inhibition of these immune inhibitory molecules is an overall activation or reactivation of the antitumor effects of endogenous cytotoxic T cells. The liver poses unique challenges to effective immunotherapy, however, as it is generally considered be relatively immune tolerant given its exposure to a high number of antigens from the gut. For example, several liver-specific non-parenchymal cells such as liver sinusoidal endothelial cells and Kupffer cells (KCs) release IL-10, which leads to T cell tolerance rather than immunity (57). Therefore, insight into the liver immune landscape may unveil novel therapeutic strategies, and help identify patients that may benefit the most from immune-modulation.
An additional factor to consider in the immune milieu in HCC is that the chronic inflammation from viral hepatitis or alcohol that predisposes to these cancers leads to an overall environment of self-tolerance within tumor and surrounding liver. In analyses of the immune tumor microenvironment in HCC, an immunosuppressive phenotype has been demonstrated by high expression of IL-10 and low expression of inflammatory molecules such as granzyme B (58), as well as an increased clonal population of infiltrating regulatory and exhausted CD8 T cells (59). Several studies have shown higher levels of Th2-like cytokines, such as IL-4, IL-8, IL-10, and IL-15, in HCCs that are more aggressive or metastatic (60), and elevated serum IL-10 levels have been associated with poorer prognosis after resection (61). By contrast, longer patient survival correlates with higher densities of infiltrating CD3+ and CD8+ cells in tumors (62), and those in which tumor infiltrating lymphocytes express a higher level of pro-inflammatory genes, such as TNF, IL-6, and CCL2 (63). While CD8+ T cells recognizing tumor-associated antigens are found in the peripheral blood of patients with HCC, their number and ability to produce IFNγ has been shown to be reduced within the tumor itself, which also contains regulatory T cells (64). The presence of greater numbers of regulatory T cells also correlates with higher TNM stage and poorer survival (65,66). In addition to high levels of immunosuppressive cytokines and cell types, several studies have demonstrated the presence of immune checkpoint inhibitors and their effects on clinical outcome in HCC. CD8+ T cells expressing PD-1 are present within HCCs, and the number of PD-1 positive CD8+ cells correlates with disease progression and post-operative; analogous findings have been shown for high PD-L1 expressing tumors (67).
Several strategies aimed at overcoming immune tolerance in the liver to enhance the immune response to HCC have been evaluated in clinical trials. Alpha-fetoprotein (AFP) was initially investigated as a potential antigen for vaccine therapy, but early results were unimpressive and the few partial responses were short (68). A vaccine against peptide components of glypican-3 (GPC3), a proteoglycan that is frequently overexpressed on the cell membrane in HCC, was found to be safe in a phase I trial and increased CD8+ T cell infiltration in tumors in most patients. Overall survival was longer in patients who developed increased GPC3 specific T cells, but only 1 of 33 patients exhibited an objective tumor response (69). A vaccine against the telomerase-derived peptide GV1001 did lead to any anti-tumor effects (70). Several small prospective studies have evaluated dendritic cell (rather than peptide) based vaccines; while these vaccines appear to be safe and can generate immune responses, no objective tumor responses have been observed in most studies (67).
Adoptive cellular therapy (ACT) is an alternative immunotherapeutic approach involving the removal of T cells from a patient and manipulating them in vitro prior to returning them to the patient. Cytokine induced killer (CIK) therapy involves isolating peripheral blood mononuclear cells (PBMCs) from a patient and activating the autologous lymphocytes with IL-2; this technique has been studied in Asia with several prospective and retrospective studies. Two randomized controlled trials showed improvement in recurrence-free survival, but no difference in overall survival when adjuvant CIK was following HCC resection (71,72). A third randomized controlled trial did demonstrate an overall survival benefit of CIK when given as adjuvant treatment in conjunction with radiofrequency ablation or transarterial chemoembolization as local control for patients who were not surgical candidates (73). Other immunomodulatory strategies are further from clinical application but are potentially promising. Chimeric antigen receptor (CAR) T cells are engineered to express specific T cell receptors that recognize pre-defined tumor antigens, and when given back to the patient should elicit an effective anti-tumor response. CAR T cells against GPC3 have shown promise in their ability to reduce HCC tumor volume in mouse models (74), but have not yet been used in human PLCs.
On September 22, 2017 the FDA granted accelerated approval to nivolumab, an anti-PD-1 monoclonal antibody, for HCC patients who failed first line sorafenib based on the CheckMate 040 trial (75). In this phase I/II non-comparative, multicenter, open-label study of 202 HCC patients with Childs A cirrhosis, nivolumab had a manageable safety profile, 20% of patients had an objective response, and 3 patients achieved a complete response. Ongoing phase III trials are needed to confirm these findings. Tremelimumab, a blocking antibody against CTLA-4, which is highly expressed on regulatory T cells, was studied in a clinical trial including 17 patients with hepatitis C related HCCs that were not amenable to other therapy. Similar to the results seen with nivolumab, 17% of patients had an objective remission and 76% had disease control (76), providing further support for the role of immune checkpoint inhibition in HCC treatment. When tremelimumab was evaluated in combination with radiofrequency ablation or chemo-ablation, 26% of patients achieved partial responses outside of the ablation zone with the addition of CTLA-4 blockade; in those patients the number of activated CD4+ and CD8+ cells in PBMCs also increased (77). The next steps in refining our immunotherapeutic strategy for HCC include evaluation of combination therapy, such as the ongoing trial of anti-CTLA-4 and anti-PD-1 therapy, and the identification and utilization of biomarkers to predict positive responses to specific treatments. Once again, the availability of models such as the TSC may aid in the discovery of clinically effective protocols.
Summary
Parallel advances in genome science, molecularly targeted- and immuno-therapies have revolutionized our approach to treating traditionally chemoresistant cancers such as HCC. This new era of molecular medicine brings hope and precision to personalized care; however, the current state of information can be overwhelming. Complex genomic data cannot be directly translated to the bedside without analyzing them in the appropriate biologic framework and providing evidence to support their functional relevance. The strategies presented here aim to bridge the existing gaps between molecular profiles, immune landscape, and meaningful therapies for patients.
Acknowledgements
Funding: Members of the Northwest Liver Research Program are supported by grants from National Institutes of Health—National Cancer Institute (CA201867: KJR), Department of Defense (CA150370: VGP, RSY), Fibrolamellar Cancer Foundation (RSY, KJR), and the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Disclaimer: Part of the article was presented at the 4th Hong Kong International Oncology Symposium 2017.
References
- Collaboration GBoDLC. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol 2017;3:1683-91. [Crossref] [PubMed]
- Bachofner JA, Valli PV, Kroger A, et al. Direct antiviral agent treatment of chronic hepatitis C results in rapid regression of transient elastography and fibrosis markers fibrosis-4 score and aspartate aminotransferase-platelet ratio index. Liver Int 2017;37:369-76. [Crossref] [PubMed]
- Saab S, Le L, Saggi S, et al. Towards the Elimination of Hepatitis C in the United States. Hepatology 2018;67:2449-59. [Crossref] [PubMed]
- Conti F, Buonfiglioli F, Scuteri A, et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J Hepatol 2016;65:727-33. [Crossref] [PubMed]
- El Kassas M, Funk AL, Salaheldin M, et al. Increased recurrence rates of hepatocellular carcinoma after DAA therapy in a hepatitis C-infected Egyptian cohort: A comparative analysis. J Viral Hepat 2018;25:623-30. [Crossref] [PubMed]
- Reig M, Marino Z, Perello C, et al. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol 2016;65:719-26. [Crossref] [PubMed]
- Kawaguchi T, Ide T, Koga H, et al. Rapidly growing hepatocellular carcinoma after direct-acting antiviral treatment of chronic hepatitis C. Clin J Gastroenterol 2018;11:69-74. [Crossref] [PubMed]
- ANRS. Lack of evidence of an effect of direct-acting antivirals on the recurrence of hepatocellular carcinoma: Data from three ANRS cohorts. J Hepatol 2016;65:734-40. [Crossref] [PubMed]
- Bielen R, Moreno C, Van Vlierberghe H, et al. The risk of early occurrence and recurrence of hepatocellular carcinoma in hepatitis C-infected patients treated with direct-acting antivirals with and without pegylated interferon: A Belgian experience. J Viral Hepat 2017;24:976-81. [Crossref] [PubMed]
- Kanwal F, Kramer J, Asch SM, et al. Risk of Hepatocellular Cancer in HCV Patients Treated With Direct-Acting Antiviral Agents. Gastroenterology 2017;153:996-1005.e1. [Crossref] [PubMed]
- Li DK, Ren Y, Fierer DS, et al. The short-term incidence of hepatocellular carcinoma is not increased after hepatitis C treatment with direct-acting antivirals: An ERCHIVES study. Hepatology 2018;67:2244-53. [Crossref] [PubMed]
- Wongjarupong N, Assavapongpaiboon B, Susantitaphong P, et al. Non-alcoholic fatty liver disease as a risk factor for cholangiocarcinoma: a systematic review and meta-analysis. BMC Gastroenterol 2017;17:149. [Crossref] [PubMed]
- Cholankeril G, Patel R, Khurana S, et al. Hepatocellular carcinoma in non-alcoholic steatohepatitis: Current knowledge and implications for management. World J Hepatol 2017;9:533-43. [Crossref] [PubMed]
- Younossi ZM, Loomba R, Rinella ME, et al. Current and Future Therapeutic Regimens for Non-alcoholic Fatty Liver Disease (NAFLD) and Non-alcoholic Steatohepatitis (NASH). Hepatology 2017. [Epub ahead of print].
- Trojan J, Waidmann O. Role of regorafenib as second-line therapy and landscape of investigational treatment options in advanced hepatocellular carcinoma. J Hepatocell Carcinoma 2016;3:31-6. [Crossref] [PubMed]
- Sia D, Losic B, Moeini A, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun 2015;6:6087. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Ho DW, Kai AK, Ng IO. TCGA whole-transcriptome sequencing data reveals significantly dysregulated genes and signaling pathways in hepatocellular carcinoma. Front Med 2015;9:322-30. [Crossref] [PubMed]
- Yu CB, Zhu LY, Wang YG, et al. Systemic transcriptome analysis of hepatocellular carcinoma. Tumour Biol 2016;37:13323-31. [Crossref] [PubMed]
- Zhang Y, Qiu Z, Wei L, et al. Integrated analysis of mutation data from various sources identifies key genes and signaling pathways in hepatocellular carcinoma. PLoS One 2014;9. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41 e23.
- Sauri T, Macarulla T, Cabrera G, et al. Comprehensive profiling of biliary tract cancers (BTC) to reveal molecular heterogeneity with implications for matched targeted therapies (MTT). J Clin Oncol 2016;34:4085.
- Verlingue L, Malka D, Allorant A, et al. Precision medicine for patients with advanced biliary tract cancers: An effective strategy within the prospective MOSCATO-01 trial. Eur J Cancer 2017;87:122-30. [Crossref] [PubMed]
- Moeini A, Sia D, Bardeesy N, et al. Molecular Pathogenesis and Targeted Therapies for Intrahepatic Cholangiocarcinoma. Clin Cancer Res 2016;22:291-300. [Crossref] [PubMed]
- Cheng AL, Finn RS, Qin S, et al. Phase III trial of lenvatinib (LEN) vs sorafenib (SOR) in first-line treatment of patients (pts) with unresectable hepatocellular carcinoma (uHCC). J Clin Oncol 2017;35:4001.
- Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56-66. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- Torrecilla S, Llovet JM. New molecular therapies for hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2015;39 Suppl 1:S80-5. [Crossref] [PubMed]
- Honeyman JN, Simon EP, Robine N, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014;343:1010-4. [Crossref] [PubMed]
- Cornella H, Alsinet C, Sayols S, et al. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology 2015;148:806-18.e10. [Crossref] [PubMed]
- Xu L, Hazard FK, Zmoos AF, et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum Mol Genet 2015;24:50-63. [Crossref] [PubMed]
- Darcy DG, Chiaroni-Clarke R, Murphy JM, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget 2015;6:755-70. [Crossref] [PubMed]
- Engelholm LH, Riaz A, Serra D, et al. CRISPR/Cas9 Engineering of Adult Mouse Liver Demonstrates That the Dnajb1-Prkaca Gene Fusion Is Sufficient to Induce Tumors Resembling Fibrolamellar Hepatocellular Carcinoma. Gastroenterology 2017;153:1662-73.e10. [Crossref] [PubMed]
- Kastenhuber ER, Lalazar G, Houlihan SL, et al. DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci U S A 2017;114:13076-84. [Crossref] [PubMed]
- Eggert T, McGlynn KA, Duffy A, et al. Epidemiology of fibrolamellar hepatocellular carcinoma in the USA, 2000-10. Gut 2013;62:1667-8. [Crossref] [PubMed]
- Riggle KM, Turnham R, Scott JD, et al. Fibrolamellar Hepatocellular Carcinoma: Mechanistic Distinction From Adult Hepatocellular Carcinoma. Pediatr Blood Cancer 2016;63:1163-7. [Crossref] [PubMed]
- Kakar S, Burgart LJ, Batts KP, et al. Clinicopathologic features and survival in fibrolamellar carcinoma: comparison with conventional hepatocellular carcinoma with and without cirrhosis. Mod Pathol 2005;18:1417-23. [Crossref] [PubMed]
- Cheung J, Ginter C, Cassidy M, et al. Structural insights into mis-regulation of protein kinase A in human tumors. Proc Natl Acad Sci U S A 2015;112:1374-9. [Crossref] [PubMed]
- Riggle KM, Riehle KJ, Kenerson HL, et al. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr Res 2016;80:110-8. [Crossref] [PubMed]
- Palechor-Ceron N, Suprynowicz FA, Upadhyay G, et al. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am J Pathol 2013;183:1862-70. [Crossref] [PubMed]
- Dilley RJ, Schwartz SM. Vascular remodeling in the growth hormone transgenic mouse. Circ Res 1989;65:1233-40. [Crossref] [PubMed]
- Broutier L, Mastrogiovanni G, Verstegen MM, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23:1424-35. [Crossref] [PubMed]
- Pauli C, Hopkins BD, Prandi D, et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov 2017;7:462-77. [Crossref] [PubMed]
- Zhang XF, Dong M, Pan YH, et al. Expression pattern of cancer-associated fibroblast and its clinical relevance in intrahepatic cholangiocarcinoma. Hum Pathol 2017;65:92-100. [Crossref] [PubMed]
- Blomme A, Van Simaeys G, Doumont G, et al. Murine stroma adopts a human-like metabolic phenotype in the PDX model of colorectal cancer and liver metastases. Oncogene 2018;37:1237-50. [Crossref] [PubMed]
- Oh BY, Lee WY, Jung S, et al. Correlation between tumor engraftment in patient-derived xenograft models and clinical outcomes in colorectal cancer patients. Oncotarget 2015;6:16059-68. [Crossref] [PubMed]
- Pergolini I, Morales-Oyarvide V, Mino-Kenudson M, et al. Tumor engraftment in patient-derived xenografts of pancreatic ductal adenocarcinoma is associated with adverse clinicopathological features and poor survival. PLoS One 2017;12. [Crossref] [PubMed]
- Gu Q, Zhang B, Sun H, et al. Genomic characterization of a large panel of patient-derived hepatocellular carcinoma xenograft tumor models for preclinical development. Oncotarget 2015;6:20160-76. [Crossref] [PubMed]
- Lancaster R. Measurement of the rate of acetylcholine diffusion through a brain slice and its significance in studies of the cellular distribution of acetylcholinesterase. J Neurochem 1971;18:2329-34. [Crossref] [PubMed]
- Schwartzkroin PA, Prince DA. Cellular and field potential properties of epileptogenic hippocampal slices. Brain Res 1978;147:117-30. [Crossref] [PubMed]
- Majumder B, Baraneedharan U, Thiyagarajan S, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun 2015;6:6169. [Crossref] [PubMed]
- Jiang X, Seo YD, Chang JH, et al. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology 2017;6. [Crossref] [PubMed]
- Mitchison NA. Studies on the immunological response to foreign tumor transplants in the mouse. I. The role of lymph node cells in conferring immunity by adoptive transfer. J Exp Med 1955;102:157-77. [Crossref] [PubMed]
- FDA approves first cancer treatment for any solid tumor with a specific genetic feature. Available online: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm
- Hato T, Goyal L, Greten TF, et al. Immune checkpoint blockade in hepatocellular carcinoma: current progress and future directions. Hepatology 2014;60:1776-82. [Crossref] [PubMed]
- Tiegs G, Lohse AW. Immune tolerance: what is unique about the liver. J Autoimmun 2010;34:1-6. [Crossref] [PubMed]
- Chew V, Lai L, Pan L, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci U S A 2017;114:E5900-E5909. [Crossref] [PubMed]
- Zheng C, Zheng L, Yoo JK, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342-56.e16. [Crossref] [PubMed]
- Hernandez-Gea V, Toffanin S, Friedman SL, et al. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 2013;144:512-27. [Crossref] [PubMed]
- Chau GY, Wu CW, Lui WY, et al. Serum Interleukin-10 But Not Interleukin-6 Is Related to Clinical Outcome in Patients With Resectable Hepatocellular Carcinoma. Ann Surg 2000;231:552-8. [Crossref] [PubMed]
- Gabrielson A, Wu Y, Wang H, et al. Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol Res 2016;4:419-30. [Crossref] [PubMed]
- Chew V, Tow C, Teo M, et al. Inflammatory tumour microenvironment is associated with superior survival in hepatocellular carcinoma patients. J Hepatol 2010;52:370-9. [Crossref] [PubMed]
- Flecken T, Schmidt N, Hild S, et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2014;59:1415-26. [Crossref] [PubMed]
- Fu J, Xu D, Liu Z, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007;132:2328-39. [Crossref] [PubMed]
- Shen X, Li N, Li H, et al. Increased prevalence of regulatory T cells in the tumor microenvironment and its correlation with TNM stage of hepatocellular carcinoma. J Cancer Res Clin Oncol 2010;136:1745-54. [Crossref] [PubMed]
- Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2015;12:681-700. [Crossref] [PubMed]
- Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, et al. The yin and yang of evasion and immune activation in HCC. J Hepatol 2015;62:1420-9. [Crossref] [PubMed]
- Sawada Y, Yoshikawa T, Nobuoka D, et al. Phase I trial of a glypican-3-derived peptide vaccine for advanced hepatocellular carcinoma: immunologic evidence and potential for improving overall survival. Clin Cancer Res 2012;18:3686-96. [Crossref] [PubMed]
- Greten TF, Forner A, Korangy F, et al. A phase II open label trial evaluating safety and efficacy of a telomerase peptide vaccination in patients with advanced hepatocellular carcinoma. BMC Cancer 2010;10:209. [Crossref] [PubMed]
- Takayama T, Sekine T, Makuuchi M, et al. Adoptive immunotherapy to lower postsurgical recurrence rates of hepatocellular carcinoma: a randomised trial. Lancet 2000;356:802-7. [Crossref] [PubMed]
- Hui D, Qiang L, Jian W, et al. A randomized, controlled trial of postoperative adjuvant cytokine-induced killer cells immunotherapy after radical resection of hepatocellular carcinoma. Dig Liver Dis 2009;41:36-41. [Crossref] [PubMed]
- Yu X, Zhao H, Liu L, et al. A randomized phase II study of autologous cytokine-induced killer cells in treatment of hepatocellular carcinoma. J Clin Immunol 2014;34:194-203. [Crossref] [PubMed]
- Ma W, Wu L, Zhou F, et al. T Cell-Associated Immunotherapy for Hepatocellular Carcinoma. Cell Physiol Biochem 2017;41:609-22. [Crossref] [PubMed]
- El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492-502. [Crossref] [PubMed]
- Sangro B, Gomez-Martin C, de la Mata M, et al. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol 2013;59:81-8. [Crossref] [PubMed]
- Duffy AG, Ulahannan SV, Makorova-Rusher O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol 2017;66:545-51. [Crossref] [PubMed]


