Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer
Introduction
The discovery of DNA structure, defined the central dogma of molecular biology with a widely acceptance in the scientific community. As far as central dogma of biology is concerned, the genetic information flows from DNA to RNA to protein. RNAs are molecules that have some similarity to DNA not only as far as structure is concerned but also in chemical composition. Transcription is the transfer of genetic code from DNA to RNA and takes place in the nucleus. RNAs then exit the nucleus into the cell body. mRNAs undergo translation in the cell body, which is the making of protein based on the code in the mRNAs. The last ones have the ability to carry the code for making proteins, so they also called “coding RNAs”. For many decades it was widely spread the idea that the majority of RNAs in our cells are mRNAs. However, recent studies have changed this “status”, suggesting instead that most RNAs do not code for proteins, and these non-coding RNAs (ncRNAs) might even hold the key to playing a great role or to furthering our understanding of human diseases. Understanding regulatory ncRNA is currently one of science’s most important challenges. Small non-coding RNAs (sncRNAs) retain a prospective role in addition to controlling the expression of most of our gene. Long non-coding RNA (lncRNA) represent the majority of transcription products, yet we know next to nothing of their significance. During the last decade, a tremendous number of ncRNAs, has been raised from anonymity in order to define as a category of genetic elements, leaving its sign on the field of tumor biology. Only a small rate of the human genome corresponds to protein-coding genes. The recent discovery, that most of our transcriptome is non-coding, is very promising. The comprehension of the biological role of this unknown RNA world undoubtedly represents the next great frontier in biology. Over the last 20 years, the intensified study of the human genome became the catalyst for a significant shift in our understanding about the way DNA operates. Complete sequencing of the human genome, within the framework of Human Genome Project (HGP), contributed to the identification and mapping of human genes. Moreover, the aforementioned international scientific consortium revealed that only a small portion, almost 2% of total DNA, is translated into proteins. As result, for a long time scientific community was holding fast to a “gene-centric” belief characterizing the majority (98%) of human DNA as “junk”. Reconsideration of the term “junk”, which was describing the non-coding part of the human genome, accomplished mainly through the results of Encyclopedia of DNA Elements project (ENCODE). This project provided with evidence supporting that most of the considered as “junk” DNA was pervasively transcribed and it could participate in the regulation of protein-coding genes by forming complex regulatory networks. Thus, the point of interest was relocated from genes to transcripts as the fundamental units of the genome (1).
ncRNAs are mainly divided into two categories based on their length, using as cutoff the 200 nucleotides (nt) length. ncRNAs <200 nt-long are referred as sncRNAs and include miRNA, siRNA, piRNA, snoRNA, snRNA and tRFs (2). ncRNAs longer than 200 nucleotides constitute an individual class of ncRNAs known as lncRNAs (Figure 1) (3). In general, ncRNAs consist the typical RNA form in mammalian cells, encompassing abundant and functional types such as rRNA and tRNA, various small RNA types such as microRNA (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA), tRNA-derived fragments (tRF), small nucleolar RNA (snoRNA), small nuclear ribonucleic acid (snRNA) as well as lncRNA. However, the overrepresentation of ncRNAs in total RNA begs the question whether or not all these RNA molecules play a biological role or they constitute “junk RNA” (4). Simultaneously, the databases including annotated ncRNA transcripts [e.g., miRBase (5), lncRNAdb (6), NONCODE (7) etc.] are expanding constantly with novel sequences, mainly due to the technological advancement through the establishment of high-throughput next-generation (NGS) technologies (8,9). In particular, NGS has revealed many ncRNAs originating from protein-coding genes (10-16). Still, the transcription itself of several sequences and/or their identification in RNA-seq data does not establish a strong argument for them to be considered as biologically active ncRNA molecules. As result, the researchers need to implement strategies based on robust biochemical and/or evolutionary data to evaluate the putative functional role of novel ncRNA candidates (4).
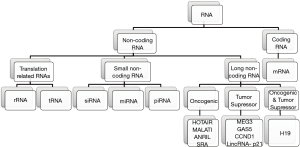
With this review, we will attempt to provide a summarizing overview of the most important classes of lncRNA and sncRNA, as they emerged from the current literature. Moreover, we will discuss in brief about their biogenesis, their implication in cellular homeostasis and cancer development as well as their potential as cancer biomarkers and therapeutic targets.
miRNAs
miRNAs are short, single-stranded, approximately 22 nucleotides in length endogenous RNA molecules. Primary miRNA (pri-miRNA) transcripts are transcribed by RNA pol II and undergo two cleavage events until mature miRNAs occur. Firstly, Drosha, an RNAse III enzyme, with its cofactor the microprocessor complex subunit DGCR8, processes pri-miRNA transcripts to precursor miRNA hairpin transcripts (pre-miRNA) in the nucleus. Then, pre-miRNA translocated to the cytoplasm via Exportin-5 (XPO5), where another RNAse III enzyme called Dicer cleaves pre-miRNA to form mature miRNA molecules that assemble into RNA-induced silencing complex (RISC) inside P-bodies (Figure 2) (17).
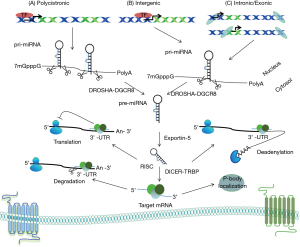
Up to date, miRNAs are the most extensively studied sncRNA molecules because of their involvement in transcriptional and post transcriptional regulation of protein-coding genes. Specifically, miRNA 5’ seed region (between nucleotides 2–7) interact with regions within the 3'untranslated region (3' UTR) of messenger RNA (mRNA) leading to the degradation or repression of the targeted mRNA(s), depending whether or not a perfect miRNA/mRNA complementarity is achieved (18). Moreover, in silico predictions suggest that over 60% of protein-coding genes could be putative targets of miRNAs. By extension, it can be declared that the expression of protein-coding genes, at least for the vast majority of them, is somehow under the regulation of miRNAs (19). Therefore, essential cellular processes as cell proliferation, cell differentiation, cell migration, angiogenesis or apoptosis are monitored by a wide, complicated miRNA network (20). As a result, any malfunction relevant to the biogenesis pathway of miRNAs is strongly associated with malignant transformation and hence renders them as key players during tumor initiation, metastasis promotion and progression of the disease (21,22).
Over the past decade, findings from several studies revealed a broad suppression of miRNAs expression in tumors compared to healthy tissues indicating for a defective miRNA-biogenesis pathway in human malignancies (23,24). A plethora of mechanisms, including genomic amplifications, deletions within fragile chromosomal sites, mutations, and epigenetic regulation of miRNA expression are responsible for the extensive deregulation of miRNA expression during carcinogenesis (25). As result, due to the fundamental mechanism of action of miRNAs to regulate specific mRNAs, they function either as oncogenes or tumor-suppressors and depending on the cellular context and different mRNA targets (26,27).
Cancer is a multistep process characterized by the ability of tumor cells to sustain chronic proliferation and continuously expand, evading growth suppressors and apoptotic signals (28-30). Numerous miRNAs have been found deregulated during oncogenesis and have been implicated in proliferation of tumor cells (31). For example, miR-17/92 cluster, which was initially proposed as an oncomiR in diffuse large B cell lymphomas (32), has been presented to be frequently overexpressed and maintain oncogenic activity in a wide spectrum of carcinomas (33). Specifically, in lymphomas, a highly notable oncogenic collaboration has been illustrated between MYC and miR-17/92. In particular, MYC is a transcriptional regulator that activates miR-17/92 cluster expression. As a result of miR-17/92 overexpression, expression of chromatin regulatory genes (SIN3B, HBP1, and BTG1) and the proapoptotic BCL2L11 gene is suppressed, contributing to survival maintenance and self-renewal of tumor cells (34). Furthermore, E2F transcription factor 1 (E2F1) is negatively regulated by miR-17/92, provoking attenuated E2F-induced apoptosis and simultaneously contributing to proliferative signal by promoting E2F transcription factor 3 (E2F3) expression (35). Members of the miR-17/92 cluster and its paralogue miR-106a/363, such as miR-92a-3p and miR-20b-5p, along with miR-155-5p have pivotal roles in other B-cell malignancies, including chronic lymphocytic leukemia (36-38). In particular, miR-155-5p regulates important transcription factors, such as E2F2 and hypoxia-inducible factor 1 (HIF1) in leukemic cells (39,40).
Hallmarks of carcinogenesis include invasion and metastasis of malignant cells as well as intense tumor angiogenesis as responding to the enormous needs for oxygen and nutrients (41). The miR-200 family members have been proposed as crucial regulators of multiple genes that are responsible for maintaining the epithelial polarity, like Zinc Finger E-Box Binding Homeobox 1 (ZEB1) and Zinc Finger E-Box Binding Homeobox 2 (ZEB2), which are controlling cadherin 1 (CDH1; also known as E-cadherin, ECAD) expression, and they actively participate in epithelial to mesenchymal transition (EMT) (42,43). Concerning the facilitation of angiogenesis by miRNAs, we could note the case of miR-126 which directly targets regulators of RAS/RAF1/MAPK pathway, crucial in angiogenic signaling [e.g., sprouty-related EVH1 domain-containing 1 (SPRED1)] (44).
Nowadays, the study of miRNA expression is mainly performed via the “gold standard” quantitative polymerase chain reaction (qPCR) followed by hybridization methodologies such as microarrays, while recent advancements in the era of NGS enabled high-throughput RNA-seq as an alternative for the in-depth analysis of miRNAs (45). As the knowledge regarding miRNAs is expanding more evidence depict the great diagnostic, prognostic and predictive potential of this novel class of biomarkers advocating for their active involvement in more and more daily clinical applications. Altered expression levels of miRNAs between malignant tumors and healthy tissues facilitate cancer diagnosis, ameliorate tumor staging and inform clinicians about relapse risk and/or disease progression as well as therapeutic efficacy, reducing the unwanted under- or overtreatment. The prognostic and predictive value improves by the synergy of more than two miRNAs, encouraging the development of multipurpose miRNA signatures as diagnostic or prognostic tools (46-48). For instance, combination of miR-15a-5p, miR-16, miR-24-3p, miR-28-5p, miR-34a, miR-96, miR-182, and miR-224, which have been proposed as molecular biomarkers with significant prognostic value in colorectal cancer (CRC); has already been proposed as a prognostic signature in this malignancy (49-57); another very important miRNA that could be added in this molecular signature is miR-21, a predictor of metastatic tumor potential in this cancer (58). Similar signatures consisting of miRNAs have been proposed in prostate cancer (59,60), laryngeal squamous cell carcinoma (61), and other human malignancies (62). Multiparametric panels of biomarkers could also integrate protein-coding transcripts produced by cancer-related genes such as the apoptosis-related genes BCL2L12 (63-67), BCL2 (68,69) and BAX (70,71), DDC (72-75), transcription factors such as HIF1 (39), and/or other mRNAs with significant prognostic value in human malignancies (76-83). Besides that, miRNAs can stably circulate in human body fluids (e.g., plasma, urine etc.), permitting their easy quantification paving the way for the development of non-invasive assays in the era of personalized medicine (84,85). Moreover, proteins that are targeted by important miRNAs could also participate in such multiparametric panels of biomarkers (86-89), along with clinical markers (90-92).
tRFs
tRFs constitute novel sncRNA molecules generated by specific cleavage of tRNA transcripts. There are two classes of tRF based on their length and their position of origin in primary or mature tRNA. The first includes stress induced tRFs, known as tRNA halves (tiRNAs or tiRs), generated by a specific cleavage by angiogenin within the anticodon loop of mature tRNAs to produce fragments ranging from 31 to 40 nucleotides. There are two subclasses of tiRNAs depending whether they include the 5’ or 3’ part of anticodon loop after cleavage (93). Interestingly, these angiogenin-produced fragments differ from other sncRNAs, including miRNAs and other tRF classes, by carrying a 5’hydroxyl group. tiRNA production by stressed cells repress translation and modulate intrinsic stress-response program of the cells. Moreover, they interact with Argonaute (AGO) protein members (e.g., AGO2) to form complexes participating in RNA interference (RNAi) silencing pathway (94). Recently, overexpression of tiRNAs in a sex hormone-dependent manner has been identified in breast cancer and prostate cancer cells positive for estrogen and androgen receptors (ARs) respectively. These sex-hormone dependent tiRNAs enhance cell proliferation, however, the mechanism of their action needs to be further investigated (95).
The second tRF class includes smaller tRNA fragments 14–30 nucleotides long, from the ends of mature or primary tRNAs which are under the spotlight of scientific community due to their size and similarity to miRNAs. Based on their mapping to 5’ or 3’ ends of tRNAs or 3’ ends of primary tRNAs are divided in three subtypes tRF-5, tRF-3 and tRF-1 respectively (96). The molecular mechanism and the enzymes that participate to the cleavage of tRNAs producing tRF-5 and tRF-3 are still unknown. At the same time, a Drosha or Dicer dependent tRNA-process mechanism has been excluded due to the maintenance of tRF abundance in experiments using Dicer and Drosha knock-out cells. Regarding their cellular distribution tRF-5 can be found in the nucleus whereas tRF-3 are predominantly gathered in the cytoplasm (97). These data are further supported by studies indicating a strong association of tRF-5 with Piwi proteins in monocytes to repress CD1A expression epigenetically through the methylation of its promoter (98). Moreover, it has been proved that tRF-3s interact with AGO proteins and act in a miRNA-like way to regulate crucial oncogenes or tumor suppressors (99).
On the contrary, tRF-1 consists a more variable tRF subclass generated by a 3’ cut of tRNA precursors from RNase Z (100). Also, tRF-1 interact with members of Piwi and AGO protein families and potentially regulate gene expression in a piRNA- or miRNA-like manner (100,101). Representatives of tRF-1 class have been found to accelerate proliferation in prostate cancer cell lines (96), or more recently to associate with the development of an aggressive CLL phenotype by regulating T cell leukemia/lymphoma 1A (TCL1A) gene (102).
Recently, tRF-2 (103) and internal tRFs (i-tRFs) (104) have been proposed as novel classes to describe other abundant tRFs. tRF-2 derived from anticodon stem loop of tRNAs and bind to Y-box binding protein 1 (YBX1) in breast cancer cells. YBX1 is a critical RNA-binding protein for the stabilization of multiple oncogenic transcripts (103). Additionally, in silico analysis of available (TCGA) datasets revealed a distinct category of tRFs, referred as i-tRFs, that spans entirely internal to mature tRNA sequences, exhibiting a cell type dependent expression and discriminating breast cancer histological subtypes (104).
piRNAs
P-element-induced wimpy testis (Piwi)-interacting RNA (piRNA) are 21–36 nucleotides single stranded RNA molecules and represent the largest group of sncRNAs. They are abundant in spermatogenic cells and they have been found to be critically involved in germline development and functions (105). In contrast to miRNA or siRNA mode of action, targeting transcripts directly, piRNAs interact with Piwi proteins, a subfamily of AGO proteins, to mediate epigenetic silencing (106). However, recent studies using Drosophila and mice as model organisms propose a potential miRNA-like function for piRNAs in the cytoplasm (107,108). piRNAs are mainly responsible for preserving genome integrity through the silencing of transposable elements (TE) (109).
Biogenesis of piRNAs has not been fully understood yet, thus several mechanisms have been proposed. The primary maturation mechanism involves the cleavage by Piwi proteins of a long primary RNA, transcribed from genomic regions identified as piRNA clusters (109). Ping-pong amplification cycle is another well-studied mechanism, characteristic of piRNAs, which bridges the gap between piRNA biogenesis and target silencing. In Drosophila germ cells a primary piRNA is associated with aubergine protein (AUB) to detect and slice active TEs. The aforementioned cleavage produces the 5' ends of new piRNA in sense with the cleaved TEs. Next, they are loaded into AGO3 and further maturate by trimming of their 3' ends. As result, new antisense piRNAs, similar to the original, are produced and target TEs (110).
piRNAs are functional RNA molecules implicated in gene silencing, thus raising the interest of scientist to define their potential role in human malignancies. Expression of oncogenes or tumor-suppressors carrying transposon-derived sequences in their 3’UTR could be modulated by piRNA (107). Deregulated piRNA fail to control and suppress the activity of TEs increasing genomic instability and promoting mutagenesis, leading to the development of aggressive cancer phenotypes (111). Moreover, Martinez et al. (112) recently revealed tissue specific piRNA expression patterns and specific piRNA-signatures between cancerous and healthy samples using The Cancer Genome Atlas (TCGA) datasets.
In breast cancer (BC) piR-36011, piR-31106 and piR-36717 have been found to differentially expressed in hormone-responsive BC, but also between cancer and normal breast tissue specimens (113). Additionally, four piRNA (piR-4987, piR-20365, piR-20485 and piR-20582) were confirmed to be upregulated in breast cancer using deep sequencing data and qPCR. Notably, piR-4987 was associated with positive lymph node status, indicating the great potential of piRNA as biomarkers in BC (114). A very recent study illustrated piR-1245 as a frequently over-expressed piRNA in CRC. Furthermore, patients with elevated piR-1245 levels were susceptible to metastasis and demonstrated lower overall survival (OS) (115). piR-651 has been proposed as a putative oncogenic piRNA as it has been found highly upregulated in gastric cancer (GC) tissues with this upregulation to be also validated in different human cancer cell lines including hepatic, cervical, breast, mesothelioma and lung (116). Similarly, piR-823 found to be upregulated in multiple myeloma (MM) patients and cell lines. Moreover, it correlated positively with the stage of the disease and characterized as a crucial molecule in the process of DNA methylation and an important regulator of myelomagenesis by stimulating bone marrow neo-angiogenesis (117). Interestingly, piR-651 and piR-823 have been used as peripheral blood biomarkers for the detection of circulating cancer cells of GC patients, able to discern patients with GC from the healthy individuals (118).
snoRNAs
snoRNAs are a distinct regulatory class of sncRNA (60–250 nucleotides) operating as guide molecules during post-transcriptional chemical modifications, such as 2’-O-methylation and pseudouridylation, of rRNA or other RNA molecules. Distinct sequence motifs and secondary structure classify snoRNA as C/D box, H/ACA box or small Cajal body-specific RNAs (scaRNA) (119). Box C/D snoRNAs (60–200 nucleotides) are distinguished by the presence of two highly conserved canonical motifs referred as C box (RUGAUGA motif, where R is a purine) and D box (CUGA motif). These snoRNAs catalyzing the site-specific 2’-O-ribose methylation of rRNA residues. Box H/ACA snoRNAs are longer than box C/D snoRNAs, ranging from 120 to 250 and guiding pseudouridylation of rRNA residues. A two-hairpin structure connected by an H box region (ANANNA, N corresponds to nucleotide) is characteristic for box H/ACA snoRNAs. Moreover, ACA trinucleotide is located three nucleotides upstream of the 3’-end of the snoRNA (120). ScaRNA are larger than the other snoRNA classes, accumulated in Cajal bodies and they characterized by the presence of both C/D and H/ACA boxes along with a CAB box (UGAG motif) that functions as a Cajal-body specific localization signal (121).
Besides their role in modification and maturation of rRNA, strong evidence has emerged concerning their potential role in human malignancies. The significance of these data is enhanced by several studies indicating a potential miRNA-like function for some small RNAs, derived from snoRNAs, after Dicer processing and interacting with AGO proteins (122,123). snoRNA U50 has been identified as a candidate tumor suppressor, downregulated in prostate and breast cancer (124,125).
Liao et al. (126) presented six deregulated snoRNAs (SNORA42, SNORD33, SNORD66, SNORD76, SNORD78, and SNORD73B) in plasma of non-small-cell lung cancer (NSCLC) patients. Of them SNORA42 was proposed to be implicated in tumorigenesis through two different pathways, a p53-dependent and a p53-independent. Notably, NSCLC patients with elevated SNORA42 expression succumbed earlier from the disease (127). In the same context, twenty-two snoRNAs were identified to demonstrate alterations specific to cancer stem cells of NSCLC patients (128). Human cell line screening of the most representative leukemic groups, including acute myeloid leukemia (AML), pre-B-acute lymphoblastic leukemia (ALL) and T-acute lymphoblastic leukemia (ALL) revealed distinct snoRNA expression patterns between various leukemic groups, however the discriminatory potential of these snoRNA, as well as their involvement in pathobiology of human leukemias, need to be further elucidated (129).
lncRNAs
Approximately 16,000 genes of the entire gene code databases belong to an important class of ncRNAs greater than 200 nucleotides in length and this number is increasing with a great rate. Very few of these RNA molecules have been characterized at all apart from their detection in different sample types. What we do know, is that expression of many of these lncRNAs is highly tissue specific and many are detected only under certain stress conditions. These fascinating molecules called lncRNAs. The last ones can be exonic, intergenic, in enhancer regions, or in regions distal to protein-coding genes. Similar to mRNAs, lncRNAs are transcribed by RNA polymerase II (RNApol II), carry single nucleotide polymorphisms (SNPs), can undergo alternative splicing, may have 5’ caps, and are usually polyadenylated. It is mentionable that under estimations the majority of lncRNAs has more than two exons, and can have secondary and tertiary structures (130). lncRNAs can function in multiple ways. They can act as scaffolds (for example NEAT1 and HOTAIR have the ability to act in trans) or guides (for example Xist, Kcnq1ot1, Airn have the ability to act in cis whereas HOTAIR acts in trans). But in addition to these two simple ostensible mechanisms, there is a plenty number of others by which lncRNAs can function and influence on cell operation. Finally, they present the ability to act as enhancers (e.g., eRNAs, in cis), reservoirs (e.g., H19) but also as decoys (e.g., Tsix, MALAT1). A tremendous increasing number of experimental studies, are providing evidence that lncRNAs mediate human disease pathogenesis, thereby challenging the concept, that protein-coding genes, are the sole contributors to the development of human disease pathogenesis. As the scientists investigate, the encoding RNA is carrying out varied cell operations in both cytoplasm and the nucleus very often involved in regulation of gene expression.
A plenty number of lncRNAs are located exclusively in the cytoplasm and others in nuclear. The last ones appeared to be heavily involved in genetic regulation of gene transcription. They recruit and guide proteins involved in modifying chromatin structure. Many lncRNAs are nuclear retained or have long residence time in the nucleus and as a consequence are often inefficient targets of RNAi. In addition, scientific studies have been descripted that a plenty number of lncRNAs are often expressed from complex loci with not only overlapping sense but also and antisense transcription (131).
lncRNAs & human malignancies
It is being recognized that certain single-nucleotide polymorphisms (SNPs) are associated with tumor risk. Large-scale data analysis from cancer genome-wide association studies indicates that the majority of SNPs associate with non-coding genes (132,133). The majority of recurrent mutations in somatic cells, copy number alterations, and tumor-related SNPs are related to ncRNAs (134-136), and the presence of risk SNPs may modulate the expression of corresponding ncRNAs. Among this number of non-coding genes, lncRNAs are emerging as a new team of indispensable members involved in the development and progression of tumor (137-140).
Moreover, the dysregulation of a number of lncRNA targets, has correlated with the prognosis and diagnosis of a plenty number of cancer types including prostate cancer (141,142), lung cancer (143), and breast cancer (144-146), among other tumor types (147,148), as well as being linked to detention against chemotherapy and targeted therapy (149-152). Correlation with a great number of analyses, indicates that these molecules are upregulated in cancer cells that are resistant to DNA damage inducers (153-156), targeted therapies or anti-hormone therapies (141,157-159). Loss-of-function studies using small hairpin RNA-based knockdown and clustered regularly interspaced short palindromic repeats (CRISPR)/cas9-mediated genetic depletion indicate that lncRNAs facilitate cancer cell growth, cell mobility and apoptosis detention, (150,160,161). The expression of HOTAIR activates estrogen receptor (ER) target transcription program and contributes to resistance to tamoxifen (161). Gain-of-function studies suggest that increased expression of lncRNAs enhances cell viability during drug treatment (153,158,162). lncRNA derives from several sources including the antisense strand of the protein coding sequence, intronic transcription, intergenic regions, or alternative splicing (163,164). A considerable percentage of known lncRNAs either reside within the cytosol or shuttle between the nucleus and the cytoplasm (165).
Recent studies present that these types of cytoplasmic lncRNAs play a great functional role in modulating mRNA translation and decay in a base-pairing dependent manner (166-168) or by competing with miRNA-mediated or a protein- mRNA decoy (169). In addition, cytoplasmic lncRNAs have been shown to regulate cytoplasmic protein trafficking from the cytosol to the nuclear areas for transcriptional activation (170). Current studies have also indicated that lncRNAs may associate with proteins, metabolic intermediates and cellular lipids. Although still largely unexplored, it has been suggested from scientists that lncRNAs are part of an essential intracellular signaling pathway. Novel type optical aspects into the regulatory roles of lncRNAs in tumor for governing new type mechanisms and pathways by which tumor cells acquire their metastatic and invasiveness properties serve as the basis of a new insight in the battle against tumorigenesis. This empathy of lncRNAs in tumor signaling should stimulate new directions for future research therapeutic options that focus on lncRNAs as novel tumor diagnostic, prognostic markers and therapeutic targets.
lncRNAs in prognosis & cancer therapy
A mentionable number of lncRNAs is deregulated in cancer and contribute to oncogenesis. In a plenty number of tumors, several lncRNAs as well as ncRNAs being transcribed from protein-coding genes (nonsense-mediated mRNA decay candidates) have been reported to be overexpressed and proposed as biomarkers (171-173). For example, several lncRNAs (GAS5 or H19) have been reported to be frequently or consistently overexpressed in urothelial carcinoma and have been proposed as individual diagnostic or prognostic biomarkers; some were moreover demonstrated to influence survival, proliferation, migration and other cancer-relevant properties of UC cell lines (174).
From a clinical aspect, lncRNAs serve as novel promising therapeutic targets. A plenty number of therapeutic strategies have been developed to target and to manipulate lncRNAs. Antisense oligonucleotide (ASO)-based strategies that downregulate the transcripts of lncRNAs via RNaseH-dependent degradation are under active investigation (175). Alternatively, nanoparticle-delivered siRNAs have been developed to knockdown lncRNAs in vivo via Argonaute- and Dicer-dependent RNA silencing (176-178), which have been evaluated in many types of models and have been found to inhibit tumor genesis and distant metastasis (178,179). Small-molecule inhibitors to block lncRNA-protein interactions or interfere with lncRNA-protein complex formation, are also on the rise. Interestingly, many types of tumors frequently become resistant to administered chemotherapeutic agents. In these chemotherapy-resistant cancers, dysregulated lncRNAs have the ability to contribute significantly to the development of this resistance (175). During clinical trials, combinations of pathway-specific inhibitors integrated with an lncRNA-directed strategy could provide maximum efficacy in treating human tumors, which is under active investigation.
Targeting lncRNAs using a variety of technologies, including ASO-based strategies, siRNAs and small molecular inhibitors should be evaluated for their effects on tumor initiation, progression or metastasis and response to therapy ASOs, including duplex RNA (180), ASO gapmers (179), and locked nucleic acids (LNAs) (181) present the ability of bind base pairing with lncRNA transcripts. The RNA-DNA duplex triggers RNase-H-dependent cleavage (182). The modern modified generation of ASOs incorporates chemical/biological modification of the backbone, constructed by sugar to improve in vivo stability and in binding affinity (183). S-constrained ethyl (cEt) modifications (184) and LNAs (181,185,186) have been advanced to pre-clinical experiments (187-189). The major characteristic of LNAs which are constructed nucleotides is the fact that contain an extra covalent bond between the 4’-C and 2’-O of the ribofuranose ring (190,191). The LNA-DNA-LNA gapmers have the ability to pair with RNA targets, which can be used to silence RNA targets not only in animal models but also in cell-line-based experiments. A similar purpose as LNAs can serve by the incorporation of bridged nucleic acid (BNA) monomers (192). In order to study the effects on cancer growth and metastasis the immediate application of ASOs to knockdown lncRNAs in vivo has been tested in a plenty of tumor models (193). LNAs present the great ability to target plasmacytoma variant translocation 1 (PVT1) and this fact has as a result the sensitization of cervical cancer cells to cisplatin, substantiating the effectiveness of combinatorial treatment (194). Clinical trials or scientific experiments using LNAs targeting AR (195) or oncoprotein Bcl-2 (196), have presented a promising result and this has as a result to be under consideration. Beyond its multiple uses in tumor, LNAs have also been proposed to improve the status of patients with cardiovascular disorders (197), neuronal diseases (198), kidney disorders (199), and other human diseases. Studies have indicated that, dioleoyl phosphatidylcholine (DOPC)-based nanoliposomes have been developed and constructed in order to deliver the nucleotide based-therapeutics (miRNA, siRNA, ASOs, and lncRNA) for clinical trial (196,200-207). Experiments have presented that a single injection of DOPC-nanoliposomal siRNAs can promote or influence the expression of target proteins for four days in mice tumors (200,201). This unique administration promotes a significant repression in the levels of expression of the gene targets (for example, BCL2, KRAS, eEF2K, miR34a, miRs155, and JAK2) and in size as far as is concerned the tumors in rodent models and preclinical models of human tumors, including many kinds of tumor models (e.g., xenografts or orthotopic models) (196,200,201,203-205,208,209).
Last but not least, RNA molecules consist targets for small-molecule inhibitors. Via high-throughput screening, scientists can identify small-molecule compounds that may potentially inhibit RNAs (210-212). There are a great number of serious efforts in order to establish platforms and methods to aid the design and identification of small molecule inhibitors for oncogenic ncRNAs but with non-mentionable results (213), fact that will facilitate the huge development of clinical or pharmaceutical agents that target lncRNAs molecules.
Conclusions
Future studies on the regulatory and biological roles of sncRNAs and lncRNAs in cancer signaling will define the future of the field. Although a huge list of ncRNAs has been identified thus far, it has been a strenuous task to demonstrate the functional relevance of ncRNAs in cancer. To answer this problem, thorough examinations of ncRNAs candidates involved in cancer signaling pathway need to be conducted to reveal the physiological relevance of ncRNAs in cell apoptosis, survival, metastasis, and metabolism. Cellular and xenograft models have been the common means of studying the roles that ncRNAs play in cancer and are useful tools in cursory evaluations of their functions. However, conclusions that are more definitive will require representative in vivo models of cancer, such as genetic models that better recapitulate the tumor microenvironment. It will be crucial to determine if tissue-specific expression of ncRNAs can induce tumor formation, which can then be blocked by targeting the ncRNAs. Identification of the specific ncRNAs that function in various human cancer types has enabled the development of ncRNA-based clinical applications such as biomarkers for diagnosis, prognostic indicators, drug sensitizers, and therapeutic targets. The ncRNAs profile of each human cancer type should be systematically investigated to improve clinical outcomes for cancer patients by engendering a personalized approach to medicine.
Acknowledgements
The research presented was carried out within the framework of a Stavros Niarchos Foundation grant to the National and Kapodistrian University of Athens.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012;337:1159, 1161.
- Romano G, Veneziano D, Acunzo M, et al. Small non-coding RNA and cancer. Carcinogenesis 2017;38:485-91. [Crossref] [PubMed]
- Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol 2009;21:416-25. [Crossref] [PubMed]
- Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet 2015;6:2. [Crossref] [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140-4. [Crossref] [PubMed]
- Amaral PP, Clark MB, Gascoigne DK, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 2011;39:D146-51. [Crossref] [PubMed]
- Liu C, Bai B, Skogerbo G, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res 2005;33:D112-5. [Crossref] [PubMed]
- Mutz KO, Heilkenbrinker A, Lonne M, et al. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol 2013;24:22-30. [Crossref] [PubMed]
- Parmakelis A, Kotsakiozi P, Kontos CK, et al. The transcriptome of a "sleeping" invader: de novo assembly and annotation of the transcriptome of aestivating Cornu aspersum. BMC Genomics 2017;18:491. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Discovery of novel transcripts of the human tissue kallikrein (KLK1) and kallikrein-related peptidase 2 (KLK2) in human cancer cells, exploiting Next-Generation Sequencing technology. Genomics 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Identification and molecular cloning of novel transcripts of the human kallikrein-related peptidase 10 (KLK10) gene using next-generation sequencing. Biochem Biophys Res Commun 2017;487:776-81. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Tsiakanikas P, et al. Identification of novel alternative splice variants of the BCL2L12 gene in human cancer cells using next-generation sequencing methodology. Cancer Lett 2016;373:119-29. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Molecular cloning of novel transcripts of human kallikrein-related peptidases 5, 6, 7, 8 and 9 (KLK5 - KLK9), using Next-generation sequencing. Sci Rep 2017;7:17299. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Scorilas A. Novel splice variants of the human kallikrein-related peptidases 11 (KLK11) and 12 (KLK12), unraveled by Next-Generation Sequencing technology. Biol Chem 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Kontos CK, Scorilas A. Molecular cloning of novel alternatively spliced variants of BCL2L12, a new member of the BCL2 gene family, and their expression analysis in cancer cells. Gene 2012;505:153-66. [Crossref] [PubMed]
- Adamopoulos PG, Raptis GD, Kontos CK, et al. Discovery and expression analysis of novel transcripts of the human SR-related CTD-associated factor 1 (SCAF1) gene in human cancer cells using Next-Generation Sequencing. Gene 2018;670:155-65. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-12. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Thomson JM, Newman M, Parker JS, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006;20:2202-7. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol 2011;223:102-15. [Crossref] [PubMed]
- Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res 2010;20:589-99. [Crossref] [PubMed]
- Christodoulou MI, Kontos CK, Halabalaki M, et al. Nature promises new anticancer agents: Interplay with the apoptosis-related BCL2 gene family. Anticancer Agents Med Chem 2014;14:375-99. [Crossref] [PubMed]
- Kontos CK, Christodoulou MI, Scorilas A. Apoptosis-related BCL2-family members: Key players in chemotherapy. Anticancer Agents Med Chem 2014;14:353-74. [Crossref] [PubMed]
- Tarushi A, Raptopoulou CP, Psycharis V, et al. Copper(II) Inverse-[9-Metallacrown-3] Compounds Accommodating Nitrato or Diclofenac Ligands: Structure, Magnetism, and Biological Activity. Eur J Inorg Chem 2016;2016:219-31. [Crossref]
- Avgeris M, Stravodimos K, Scorilas A. Loss of miR-378 in prostate cancer, a common regulator of KLK2 and KLK4, correlates with aggressive disease phenotype and predicts the short-term relapse of the patients. Biol Chem 2014;395:1095-104. [Crossref] [PubMed]
- Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 2004;64:3087-95. [Crossref] [PubMed]
- Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J 2012;18:262-7. [Crossref] [PubMed]
- Dal Bo M, Bomben R, Hernandez L, et al. The MYC/miR-17-92 axis in lymphoproliferative disorders: A common pathway with therapeutic potential. Oncotarget 2015;6:19381-92. [Crossref] [PubMed]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 2007;282:2130-4. [Crossref] [PubMed]
- Papageorgiou SG, Diamantopoulos MA, Kontos CK, et al. MicroRNA-92a-3p overexpression in peripheral blood mononuclear cells is an independent predictor of prolonged overall survival of patients with chronic lymphocytic leukemia. Leuk Lymphoma 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Diamantopoulos MA, et al. MicroRNA-155-5p Overexpression in Peripheral Blood Mononuclear Cells of Chronic Lymphocytic Leukemia Patients Is a Novel, Independent Molecular Biomarker of Poor Prognosis. Dis Markers 2017;2017. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Tsiakanikas P, et al. Elevated miR-20b-5p expression in peripheral blood mononuclear cells: A novel, independent molecular biomarker of favorable prognosis in chronic lymphocytic leukemia. Leuk Res 2018;70:1-7. [Crossref] [PubMed]
- Kontos CK, Papageorgiou SG, Diamantopoulos MA, et al. mRNA overexpression of the hypoxia inducible factor 1 alpha subunit gene (HIF1A): An independent predictor of poor overall survival in chronic lymphocytic leukemia. Leuk Res 2017;53:65-73. [Crossref] [PubMed]
- Kontos CK, Vasilatou D, Papageorgiou SG, et al. Translation Regulation by microRNAs in Acute Leukemia. Reviews in Cell Biology and Molecular Medicine. Wiley-VCH Verlag GmbH & Co. KGaA, 2014.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894-907. [Crossref] [PubMed]
- Skourti E, Logotheti S, Kontos CK, et al. Progression of mouse skin carcinogenesis is associated with the orchestrated deregulation of mir-200 family members, mir-205 and their common targets. Mol Carcinog 2016;55:1229-42. [Crossref] [PubMed]
- Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261-71. [Crossref] [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [Crossref] [PubMed]
- Avgeris M, Mavridis K, Tokas T, et al. Uncovering the clinical utility of miR-143, miR-145 and miR-224 for predicting the survival of bladder cancer patients following treatment. Carcinogenesis 2015;36:528-37. [Crossref] [PubMed]
- Piatopoulou D, Avgeris M, Drakaki I, et al. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann Hematol 2018;97:1169-82. [Crossref] [PubMed]
- Tsikrika FD, Avgeris M, Levis PK, et al. miR-221/222 cluster expression improves clinical stratification of non-muscle invasive bladder cancer (TaT1) patients' risk for short-term relapse and progression. Genes Chromosomes Cancer 2018;57:150-61. [Crossref] [PubMed]
- Kerimis D, Kontos CK, Christodoulou S, et al. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin Biochem 2017;50:285-92. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Papadopoulos IN, et al. Enhanced miR-182 transcription is a predictor of poor overall survival in colorectal adenocarcinoma patients. Clin Chem Lab Med 2014;52:1217-27. [Crossref] [PubMed]
- Tsiakanikas P, Kontos CK, Kerimis D, et al. High microRNA-28-5p expression in colorectal adenocarcinoma predicts short-term relapse of node-negative patients and poor overall survival of patients with non-metastatic disease. Clin Chem Lab Med 2018;56:990-1000. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Papadopoulos IN, et al. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumour Biol 2016;37:11815-24. [Crossref] [PubMed]
- Kontos CK, Tsiakanikas P, Avgeris M, et al. miR-15a-5p, A Novel Prognostic Biomarker, Predicting Recurrent Colorectal Adenocarcinoma. Mol Diagn Ther 2017;21:453-64. [Crossref] [PubMed]
- Rapti SM, Kontos CK, Christodoulou S, et al. miR-34a overexpression predicts poor prognostic outcome in colorectal adenocarcinoma, independently of clinicopathological factors with established prognostic value. Clin Biochem 2017;50:918-24. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Rapti SM, et al. miR-224 overexpression is a strong and independent prognosticator of short-term relapse and poor overall survival in colorectal adenocarcinoma. Int J Oncol 2015;46:849-59. [Crossref] [PubMed]
- Diamantopoulos MA, Kontos CK, Kerimis D, et al. Upregulated miR-16 expression is an independent indicator of relapse and poor overall survival of colorectal adenocarcinoma patients. Clin Chem Lab Med 2017;55:737-47. [Crossref] [PubMed]
- Sterlacci W, Sioulas AD, Veits L, et al. 22-gauge core vs 22-gauge aspiration needle for endoscopic ultrasound-guided sampling of abdominal masses. World J Gastroenterol 2016;22:8820-30. [Crossref] [PubMed]
- Ferraro A, Kontos CK, Boni T, et al. Epigenetic regulation of miR-21 in colorectal cancer: ITGB4 as a novel miR-21 target and a three-gene network (miR-21-ITGBeta4-PDCD4) as predictor of metastatic tumor potential. Epigenetics 2014;9:129-41. [Crossref] [PubMed]
- Kontos CK, Adamopoulos PG, Scorilas A. Prognostic and predictive biomarkers in prostate cancer. Expert Rev Mol Diagn 2015;15:1567-76. [Crossref] [PubMed]
- Avgeris M, Stravodimos K, Fragoulis EG, et al. The loss of the tumour-suppressor miR-145 results in the shorter disease-free survival of prostate cancer patients. Br J Cancer 2013;108:2573-81. [Crossref] [PubMed]
- Kontos CK, Adamopoulos PG, Scorilas A. Molecular biomarkers of laryngeal cancer. Biomarkers in Disease: Methods, Discoveries and Applications: Biomarkers in Cancer. Netherlands: Spring, 2015:891-919.
- Vasilatou D, Papageorgiou SG, Kontsioti F, et al. Expression analysis of mir-17-5p, mir-20a and let-7a microRNAs and their target proteins in CD34+ bone marrow cells of patients with myelodysplastic syndromes. Leuk Res 2013;37:251-8. [Crossref] [PubMed]
- Fendri A, Kontos CK, Khabir A, et al. BCL2L12 is a novel biomarker for the prediction of short-term relapse in nasopharyngeal carcinoma. Mol Med 2011;17:163-71. [Crossref] [PubMed]
- Geomela PA, Kontos CK, Yiotakis I, et al. Quantitative expression analysis of the apoptosis-related gene, BCL2L12, in head and neck squamous cell carcinoma. J Oral Pathol Med 2013;42:154-61. [Crossref] [PubMed]
- Kontos CK, Papadopoulos IN, Scorilas A. Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene BCL2L12 in colon cancer. Biol Chem 2008;389:1467-75. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Pappa V, et al. The novel member of the BCL2 gene family, BCL2L12, is substantially elevated in chronic lymphocytic leukemia patients, supporting its value as a significant biomarker. Oncologist 2011;16:1280-91. [Crossref] [PubMed]
- Foutadakis S, Avgeris M, Tokas T, et al. Increased BCL2L12 expression predicts the short-term relapse of patients with TaT1 bladder cancer following transurethral resection of bladder tumors. Urol Oncol 2014;32:39.e29-36. [Crossref] [PubMed]
- Fendri A, Kontos CK, Khabir A, et al. Quantitative analysis of BCL2 mRNA expression in nasopharyngeal carcinoma: an unfavorable and independent prognostic factor. Tumour Biol 2010;31:391-9. [Crossref] [PubMed]
- Stamati L, Avgeris M, Kosmidis H, et al. Overexpression of BCL2 and BAX following BFM induction therapy predicts ch-ALL patients' poor response to treatment and short-term relapse. J Cancer Res Clin Oncol 2015;141:2023-36. [Crossref] [PubMed]
- Giotakis AI, Kontos CK, Manolopoulos LD, et al. High BAX/BCL2 mRNA ratio predicts favorable prognosis in laryngeal squamous cell carcinoma, particularly in patients with negative lymph nodes at the time of diagnosis. Clin Biochem 2016;49:890-6. [Crossref] [PubMed]
- Kontos CK, Fendri A, Khabir A, et al. Quantitative expression analysis and prognostic significance of the BCL2-associated X gene in nasopharyngeal carcinoma: a retrospective cohort study. BMC Cancer 2013;13:293. [Crossref] [PubMed]
- Geomela PA, Kontos CK, Yiotakis I, et al. L-DOPA decarboxylase mRNA expression is associated with tumor stage and size in head and neck squamous cell carcinoma: a retrospective cohort study. BMC Cancer 2012;12:484. [Crossref] [PubMed]
- Kontos CK, Papadopoulos IN, Fragoulis EG, et al. Quantitative expression analysis and prognostic significance of L-DOPA decarboxylase in colorectal adenocarcinoma. Br J Cancer 2010;102:1384-90. [Crossref] [PubMed]
- Avgeris M, Koutalellis G, Fragoulis EG, et al. Expression analysis and clinical utility of L-Dopa decarboxylase (DDC) in prostate cancer. Clin Biochem 2008;41:1140-9. [Crossref] [PubMed]
- Koutalellis G, Stravodimos K, Avgeris M, et al. L-dopa decarboxylase (DDC) gene expression is related to outcome in patients with prostate cancer. BJU Int 2012;110:E267-73. [Crossref] [PubMed]
- Psyrri A, Papageorgiou S, Liakata E, et al. Phosphatidylinositol 3'-kinase catalytic subunit alpha gene amplification contributes to the pathogenesis of mantle cell lymphoma. Clin Cancer Res 2009;15:5724-32. [Crossref] [PubMed]
- Adamopoulos PG, Kontos CK, Papageorgiou SG, et al. KLKB1 mRNA overexpression: A novel molecular biomarker for the diagnosis of chronic lymphocytic leukemia. Clin Biochem 2015;48:849-54. [Crossref] [PubMed]
- Alexopoulou DK, Kontos CK, Christodoulou S, et al. KLK11 mRNA expression predicts poor disease-free and overall survival in colorectal adenocarcinoma patients. Biomark Med 2014;8:671-85. [Crossref] [PubMed]
- Christodoulou S, Alexopoulou DK, Kontos CK, et al. Kallikrein-related peptidase-6 (KLK6) mRNA expression is an independent prognostic tissue biomarker of poor disease-free and overall survival in colorectal adenocarcinoma. Tumour Biol 2014;35:4673-85. [Crossref] [PubMed]
- Foteinou E, Kontos CK, Giotakis AI, et al. Low mRNA expression levels of kallikrein-related peptidase 4 (KLK4) predict short-term relapse in patients with laryngeal squamous cell carcinoma. Biol Chem 2014;395:1051-62. [Crossref] [PubMed]
- Kontos CK. Surrogate Prognostic Biomarkers in OSCC: The Paradigm of PA28gamma Overexpression. EBioMedicine 2015;2:784-5. [Crossref] [PubMed]
- Kontos CK, Adamopoulos PG, Papageorgiou SG, et al. mRNA overexpression of kallikrein-related peptidase 14 (KLK14) is an independent predictor of poor overall survival in chronic lymphocytic leukemia patients. Clin Chem Lab Med 2016;54:315-24. [Crossref] [PubMed]
- Kontos CK, Chantzis D, Papadopoulos IN, et al. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett 2013;330:106-12. [Crossref] [PubMed]
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087-92. [Crossref] [PubMed]
- Piatopoulou D, Avgeris M, Marmarinos A, et al. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br J Cancer 2017;117:801-12. [Crossref] [PubMed]
- Kefala M, Papageorgiou SG, Kontos CK, et al. Increased expression of phosphorylated NBS1, a key molecule of the DNA damage response machinery, is an adverse prognostic factor in patients with de novo myelodysplastic syndromes. Leuk Res 2013;37:1576-82. [Crossref] [PubMed]
- Miltiades P, Lamprianidou E, Vassilakopoulos TP, et al. The Stat3/5 Signaling Biosignature in Hematopoietic Stem/Progenitor Cells Predicts Response and Outcome in Myelodysplastic Syndrome Patients Treated with Azacitidine. Clin Cancer Res 2016;22:1958-68. [Crossref] [PubMed]
- Papageorgiou SG, Kontos CK, Foukas PG, et al. BCL2L12 protein overexpression is associated with favorable outcome in diffuse large B-cell lymphoma patients in the rituximab era. Leuk Lymphoma 2016;57:2199-203. [Crossref] [PubMed]
- Vassilakopoulou M, Avgeris M, Velcheti V, et al. Evaluation of PD-L1 Expression and Associated Tumor-Infiltrating Lymphocytes in Laryngeal Squamous Cell Carcinoma. Clin Cancer Res 2016;22:704-13. [Crossref] [PubMed]
- Mpakou VE, Ioannidou HD, Konsta E, et al. Quantitative and qualitative analysis of regulatory T cells in B cell chronic lymphocytic leukemia. Leuk Res 2017;60:74-81. [Crossref] [PubMed]
- Papageorgiou SG, Vasilatou D, Kontos CK, et al. Treatment with 5-Azacytidine improves clinical outcome in high-risk MDS patients in the 'real life' setting: A single center observational study. Hematology 2016;21:34-41. [Crossref] [PubMed]
- Papageorgiou SG, Vasilatou D, Kontos CK, et al. The prognostic value of monosomal karyotype (MK) in higher-risk patients with myelodysplastic syndromes treated with 5-Azacitidine: A retrospective analysis of the Hellenic (Greek) Myelodysplastic syndromes Study Group. Am J Hematol 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell 2009;138:215-9. [Crossref] [PubMed]
- Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011;43:613-23. [Crossref] [PubMed]
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A 2015;112:E3816-25. [Crossref] [PubMed]
- Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009;23:2639-49. [Crossref] [PubMed]
- Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci 2016;41:679-89. [Crossref] [PubMed]
- Zhang X, He X, Liu C, et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. J Immunol 2016;196:1591-603. [Crossref] [PubMed]
- Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A 2013;110:1404-9. [Crossref] [PubMed]
- Pekarsky Y, Balatti V, Palamarchuk A, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A 2016;113:5071-6. [Crossref] [PubMed]
- Haussecker D, Huang Y, Lau A, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010;16:673-95. [Crossref] [PubMed]
- Balatti V, Rizzotto L, Miller C, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2015;112:2169-74. [Crossref] [PubMed]
- Goodarzi H, Liu X, Nguyen HC, et al. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015;161:790-802. [Crossref] [PubMed]
- Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 2015;6:24797-822. [Crossref] [PubMed]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009;25:355-76. [Crossref] [PubMed]
- Huang XA, Yin H, Sweeney S, et al. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 2013;24:502-16. [Crossref] [PubMed]
- Watanabe T, Lin H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol Cell 2014;56:18-27. [Crossref] [PubMed]
- Zhang P, Kang JY, Gou LT, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 2015;25:193-207. [Crossref] [PubMed]
- Weick EM, Miska EA. piRNAs: from biogenesis to function. Development 2014;141:3458-71. [Crossref] [PubMed]
- Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci 2016;41:324-37. [Crossref] [PubMed]
- Moyano M, Stefani G. piRNA involvement in genome stability and human cancer. J Hematol Oncol 2015;8:38. [Crossref] [PubMed]
- Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep 2015;5:10423. [Crossref] [PubMed]
- Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014;5:9901-10. [Crossref] [PubMed]
- Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol 2013;15:563-8. [Crossref] [PubMed]
- Weng W, Liu N, Toiyama Y, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer 2018;17:16. [Crossref] [PubMed]
- Cheng J, Guo JM, Xiao BX, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 2011;412:1621-5. [Crossref] [PubMed]
- Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015;29:196-206. [Crossref] [PubMed]
- Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem 2011;44:1050-7. [Crossref] [PubMed]
- Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011;93:1987-92. [Crossref] [PubMed]
- Reichow SL, Hamma T, Ferre-D'Amare AR, et al. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 2007;35:1452-64. [Crossref] [PubMed]
- Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol 2004;14:335-43. [Crossref] [PubMed]
- Brameier M, Herwig A, Reinhardt R, et al. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 2011;39:675-86. [Crossref] [PubMed]
- Martens-Uzunova ES, Hoogstrate Y, Kalsbeek A, et al. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget 2015;6:17430-44. [Crossref] [PubMed]
- Dong XY, Guo P, Boyd J, et al. Implication of snoRNA U50 in human breast cancer. J Genet Genomics 2009;36:447-54. [Crossref] [PubMed]
- Dong XY, Rodriguez C, Guo P, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet 2008;17:1031-42. [Crossref] [PubMed]
- Liao J, Yu L, Mei Y, et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer 2010;9:198. [Crossref] [PubMed]
- Mei YP, Liao JP, Shen J, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012;31:2794-804. [Crossref] [PubMed]
- Mannoor K, Shen J, Liao J, et al. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer 2014;13:104. [Crossref] [PubMed]
- Teittinen KJ, Laiho A, Uusimaki A, et al. Expression of small nucleolar RNAs in leukemic cells. Cell Oncol (Dordr) 2013;36:55-63. [Crossref] [PubMed]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture 2012;2:189-99. [Crossref] [PubMed]
- Sun W, Yang Y, Xu C, et al. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet 2017;216-217:105-10. [Crossref] [PubMed]
- Chen G, Qiu C, Zhang Q, et al. Genome-wide analysis of human SNPs at long intergenic noncoding RNAs. Hum Mutat 2013;34:338-44. [Crossref] [PubMed]
- Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760-74. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer 2013;108:2419-25. [Crossref] [PubMed]
- Melton C, Reuter JA, Spacek DV, et al. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 2015;47:710-6. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Khurana E, Fu Y, Chakravarty D, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet 2016;17:93-108. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Guo H, Ahmed M, Zhang F, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet 2016;48:1142-50. [Crossref] [PubMed]
- Jin G, Sun J, Isaacs SD, et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis 2011;32:1655-9. [Crossref] [PubMed]
- Yuan H, Liu H, Liu Z, et al. A Novel Genetic Variant in Long Non-coding RNA Gene NEXN-AS1 is Associated with Risk of Lung Cancer. Sci Rep 2016;6:34234. [Crossref] [PubMed]
- Lin A, Hu Q, Li C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol 2017;19:238-51. [Crossref] [PubMed]
- Xia Z, Yan R, Duan F, et al. Genetic Polymorphisms in Long Noncoding RNA H19 Are Associated With Susceptibility to Breast Cancer in Chinese Population. Medicine (Baltimore) 2016;95. [Crossref] [PubMed]
- Xu T, Hu XX, Liu XX, et al. Association between SNPs in Long Non-coding RNAs and the Risk of Female Breast Cancer in a Chinese Population. J Cancer 2017;8:1162-9. [Crossref] [PubMed]
- Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A 2012;109:8646-51. [Crossref] [PubMed]
- Zhao X, Wei X, Zhao L, et al. The rs6983267 SNP and long non-coding RNA CARLo-5 are associated with endometrial carcinoma. Environ Mol Mutagen 2016;57:508-15. [Crossref] [PubMed]
- Askarian-Amiri ME, Leung E, Finlay G, et al. The Regulatory Role of Long Noncoding RNAs in Cancer Drug Resistance. Methods Mol Biol 2016;1395:207-27. [Crossref] [PubMed]
- Chen QN, Wei CC, Wang ZX, et al. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget 2017;8:1925-36. [PubMed]
- Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA Repair (Amst) 2016;45:25-33. [Crossref] [PubMed]
- Pan JJ, Xie XJ, Li X, et al. Long Non-coding RNAs and Drug Resistance. Asian Pac J Cancer Prev 2015;16:8067-73. [Crossref] [PubMed]
- Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene 2007;26:4877-81. [Crossref] [PubMed]
- Tsang WP, Wong TW, Cheung AH, et al. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 2007;13:890-8. [Crossref] [PubMed]
- Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics 2016;14:280-8. [Crossref] [PubMed]
- Zhang CL, Zhu KP, Shen GQ, et al. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol 2016;37:2737-48. [Crossref] [PubMed]
- Hou Z, Xu C, Xie H, et al. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One 2014;9. [Crossref] [PubMed]
- Meijer D, van Agthoven T, Bosma PT, et al. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res 2006;4:379-86. [Crossref] [PubMed]
- Silveira RA, Fachel AA, Moreira YB, et al. Protein-coding genes and long noncoding RNAs are differentially expressed in dasatinib-treated chronic myeloid leukemia patients with resistance to imatinib. Hematology 2014;19:31-41. [Crossref] [PubMed]
- Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621-9. [Crossref] [PubMed]
- Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016;35:2746-55. [Crossref] [PubMed]
- Wang Y, Zhang D, Wu K, et al. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol 2014;34:3182-93. [Crossref] [PubMed]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2001;2:919-29. [Crossref] [PubMed]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet 2015;31:239-51. [Crossref] [PubMed]
- Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics 2016;14:73-80. [Crossref] [PubMed]
- Beltran M, Puig I, Pena C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 2008;22:756-69. [Crossref] [PubMed]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011;470:284-8. [Crossref] [PubMed]
- Kim YK, Furic L, Parisien M, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 2007;26:2670-81. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69. [Crossref] [PubMed]
- Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370-81. [Crossref] [PubMed]
- Kontos CK, Mavridis K, Talieri M, et al. Kallikrein-related peptidases (KLKs) in gastrointestinal cancer: mechanistic and clinical aspects. Thromb Haemost 2013;110:450-7. [Crossref] [PubMed]
- Kontos CK, Scorilas A. Kallikrein-related peptidases (KLKs): a gene family of novel cancer biomarkers. Clin Chem Lab Med 2012;50:1877-91. [Crossref] [PubMed]
- Kontos CK, Scorilas A, Papavassiliou AG. The role of transcription factors in laboratory medicine. Clin Chem Lab Med 2013;51:1563-71. [PubMed]
- Droop J, Szarvas T, Schulz WA, et al. Diagnostic and prognostic value of long noncoding RNAs as biomarkers in urothelial carcinoma. PLoS One 2017;12. [Crossref] [PubMed]
- Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov 2017;16:167-79. [Crossref] [PubMed]
- Buyens K, De Smedt SC, Braeckmans K, et al. Liposome based systems for systemic siRNA delivery: stability in blood sets the requirements for optimal carrier design. J Control Release 2012;158:362-70. [Crossref] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009;136:642-55. [Crossref] [PubMed]
- Lee JM, Yoon TJ, Cho YS. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. Biomed Res Int 2013;2013. [PubMed]
- Young SW, Stenzel M, Yang JL. Nanoparticle-siRNA: A potential cancer therapy? Crit Rev Oncol Hematol 2016;98:159-69. [Crossref] [PubMed]
- Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 2012;11:125-40. [Crossref] [PubMed]
- Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 2004;43:13233-41. [Crossref] [PubMed]
- Chan JH, Lim S, Wong WS. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol 2006;33:533-40. [Crossref] [PubMed]
- Geary RS, Norris D, Yu R, et al. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 2015;87:46-51. [Crossref] [PubMed]
- Seth PP, Siwkowski A, Allerson CR, et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp Ser (Oxf) 2008:553-4.
- Moreno PM, Pego AP. Therapeutic antisense oligonucleotides against cancer: hurdling to the clinic. Front Chem 2014;2:87. [Crossref] [PubMed]
- Sarma K, Levasseur P, Aristarkhov A, et al. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A 2010;107:22196-201. [Crossref] [PubMed]
- Dürig J, Duhrsen U, Klein-Hitpass L, et al. The novel antisense Bcl-2 inhibitor SPC2996 causes rapid leukemic cell clearance and immune activation in chronic lymphocytic leukemia. Leukemia 2011;25:638-47. [Crossref] [PubMed]
- Hong D, Kurzrock R, Kim Y, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 2015;7. [Crossref] [PubMed]
- Pandey SK, Wheeler TM, Justice SL, et al. Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J Pharmacol Exp Ther 2015;355:329-40. [Crossref] [PubMed]
- Kumar R, Singh SK, Koshkin AA, et al. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2'-thio-LNA. Bioorg Med Chem Lett 1998;8:2219-22. [Crossref] [PubMed]
- Tereshko V, Teplova M, Brunzelle J, et al. Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression. Nat Struct Biol 2001;8:899-907. [Crossref] [PubMed]
- Sekiguchi M, Obika S, Somjing R, et al. Synthesis and properties of a novel bridged nucleic acid analogue, 5'-amino-3',5'-BNA. Nucleosides Nucleotides Nucleic Acids 2005;24:1097-100. [Crossref] [PubMed]
- Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180-9. [Crossref] [PubMed]
- Iden M, Fye S, Li K, et al. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS One 2016;11. [Crossref] [PubMed]
- Bianchini D, Omlin A, Pezaro C, et al. First-in-human Phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br J Cancer 2013;109:2579-86. [Crossref] [PubMed]
- Tekedereli I, Alpay SN, Akar U, et al. Therapeutic Silencing of Bcl-2 by Systemically Administered siRNA Nanotherapeutics Inhibits Tumor Growth by Autophagy and Apoptosis and Enhances the Efficacy of Chemotherapy in Orthotopic Xenograft Models of ER (-) and ER (+) Breast Cancer. Mol Ther Nucleic Acids 2013;2. [Crossref] [PubMed]
- Ottaviani L, da Costa Martins PA. Non-coding RNAs in cardiac hypertrophy. J Physiol 2017;595:4037-50. [Crossref] [PubMed]
- Khorkova O, Wahlestedt C. Oligonucleotide therapies for disorders of the nervous system. Nat Biotechnol 2017;35:249-63. [Crossref] [PubMed]
- Yang C, Zhang C, Zhao Z, et al. Fighting against kidney diseases with small interfering RNA: opportunities and challenges. J Transl Med 2015;13:39. [Crossref] [PubMed]
- Aslan B, Monroig P, Hsu MC, et al. The ZNF304-integrin axis protects against anoikis in cancer. Nat Commun 2015;6:7351. [Crossref] [PubMed]
- Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 2012;26:338-43. [Crossref] [PubMed]
- Naina HV, Harris S. Paraneoplastic thrombocytosis in ovarian cancer. N Engl J Med 2012;366:1840; author reply 1840.
- Nick AM, Stone RL, Armaiz-Pena G, et al. Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J Natl Cancer Inst 2011;103:1596-612. [Crossref] [PubMed]
- Nishimura M, Jung EJ, Shah MY, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov 2013;3:1302-15. [Crossref] [PubMed]
- Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev 2014;66:110-6. [Crossref] [PubMed]
- Pradeep S, Huang J, Mora EM, et al. Erythropoietin Stimulates Tumor Growth via EphB4. Cancer Cell 2015;28:610-22. [Crossref] [PubMed]
- Tekedereli I, Alpay SN, Tavares CD, et al. Targeted silencing of elongation factor 2 kinase suppresses growth and sensitizes tumors to doxorubicin in an orthotopic model of breast cancer. PLoS One 2012;7. [Crossref] [PubMed]
- Ozpolat B, Sood AK, Lopez-Berestein G. Nanomedicine based approaches for the delivery of siRNA in cancer. J Intern Med 2010;267:44-53. [Crossref] [PubMed]
- Rivas E, Clements J, Eddy SR. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 2017;14:45-8. [Crossref] [PubMed]
- Connelly CM, Moon MH, Schneekloth JS Jr. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem Biol 2016;23:1077-90. [Crossref] [PubMed]
- Howe JA, Wang H, Fischmann TO, et al. Selective small-molecule inhibition of an RNA structural element. Nature 2015;526:672-7. [Crossref] [PubMed]
- Velagapudi SP, Cameron MD, Haga CL, et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A 2016;113:5898-903. [Crossref] [PubMed]
- Velagapudi SP, Luo Y, Tran T, et al. Defining RNA-Small Molecule Affinity Landscapes Enables Design of a Small Molecule Inhibitor of an Oncogenic Noncoding RNA. ACS Cent Sci 2017;3:205-16. [Crossref] [PubMed]


