Increasing clinical resistance rate of Shigella sonnei to cefotaxime in Jiangsu Province, China, between 2012 and 2015
Introduction
Shigellosis is the most common communicable disorder of the gastrointestinal tract, mainly spread by fecal-to-oral transmission (1). There is an estimated annual prevalence of 165 million cases of shigellosis worldwide, with 163.2 million in the developing world and the remaining 1.5 million in industrialized countries (2). So far shigellosis has been a peril to public health, particularly in developing countries. Shigella, comprising four serotypes: S. flexneri, S. sonnei, S. boydii, and S. dysenteriae, is one of the most dominant etiological agents of bacillary dysentery. Statistical analyses demonstrated that most S. sonnei strains were detected in high-income regions, with a percentage of 77% in industrialized countries and 15% in developing countries, respectively (2-4). The proportional increase of S. sonnei infections in developing countries is becoming a global concern (5).
Cefotaxime, a third-generation cephalosporin, was initially approved for a variety of clinical bacterial pathogens such as Shigella (6) in EU and North America. Thereafter the widespread use of cefotaxime for Shigella, especially S. sonnei, inevitably resulted in bacterial resistance, as illustrated in the contrast between a null prevalence rate of resistant S. sonnei in Santiago, Chile and 88.9% in Iran (7,8), thus exacerbating the dilemma in antibiotic prescribing. Herein, our study investigated the drug-resistant patterns of S. sonnei to cefotaxime in the Jiangsu Province, China.
The emergence of cefotaxime-resistant S. sonnei was attributed to extended-spectrum beta-lactamases (ESBLs)-producing genes, encoded by plasmids which are responsible for the spread of drug resistance between bacteria, from resistant to susceptible strains via a series of procedures, i.e., conjugation, transformation, and transduction. Ever since the first description of ESBLs in Germany in 1983 (9), ESBLs-resistant genes in Shigella were identified in South Korea (10), Iran (8), Vietnam, etc. (11). Continuous emergence of ESBLs gene alerts us to the severity of expanded-spectrum cephalosporins.
In this study, we investigated and evaluated the resistance rate to cefotaxime as well as the resistance mechanism, spreading pathway of ESBL-producing clinical S. sonnei strains isolated between 2012 and 2015 in Jiangsu Province, China, which would provide the reference directing clinical prophylaxis and therapeutics.
Methods
Isolation and identification of Shigella strains
Clinical Shigella strains, collected from stools of patients from hospitals in major cities in Jiangsu Province, China. For stool testing, fresh feces with pus, blood and mucous should be collected and cultured, with each sample about 1–5 g. In the case of infants or incontinence, rectum swab was applied. Fecal samples were identified by API 20E test strips (bioMérieuxVitek, Marcy l’Étoile, France) and serotyped by slide agglutination with a commercial antiserum kit (Tianrun Bio-Pharmaceutical Co. Ltd., China).
Antimicrobial susceptibility test
Antimicrobial susceptibility tests to S. sonnei were performed by the Kirby-Bauer disk diffusion method with the following antimicrobials: cefalotin, cefotaxime, ampicillin, amoxicillin/clavulanic acid, gentamycin, nalidixic acid, norfloxacin, tetracycline, sulfamethoxazole according to the Clinical and Laboratory Standards Institute (CLSI) standards (12). The control strains were E. coli ATCC 25922 and E. coli ATCC 35218.
Detection of ESBLs genes
Genomic DNA of each S. sonnei isolate was extracted using an extraction kit (Biospin plasmid extraction, Bioflux), with genes blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, and blaOXA detected using two pairs of primes as previously reported (13). Amplification of the antibiotic-resistant genes was performed at following temperature conditions: pre-denaturation at 95 °C for 5 min, followed by 30 cycles at 95 °C for 30 s, annealing for 30 s and at 72 °C for 1 min, with a final extension procedure at 72 °C for 5 min. Polymerase chain reaction (PCR) products were analyzed by electrophoresis with 2.0% agarose.
DNA sequence analysis
PCR products with positive results for electrophoresis were sequenced and compared with the sequences of GenBank database to further identify the subtypes of the β-lactamase genes. Similarity and alignment searches for the nucleotide sequences were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST).
Results
Distribution of pathogen
A total of 95 strains of S. sonnei were isolated from hospitals in various regions in Jiangsu Province between 2012 and 2015. There were 41 strains of S. sonnei between 2012 and 2013, and 54 isolates during 2014 and 2015. Of the strains, 49 isolates were from male patients and 46 in females. In addition, 31 (32.6%) of patients with dysentery were aged 1–10, 26 (27.4%) aged 11–30, 24 (25.3%) aged 31–50 and 14 (14.7%) aged >50. With respect to cities in Jiangsu Province, Zhenjiang (24.2%, n=23) contributed to the largest proportion of S. sonnei collected, followed by Wuxi (13.7%, n=13), Changzhou (12.6%, n=12), Suzhou (9.5%, n=9), Nantong (9.5%, n=9), Nanjing (8.4%, n=8), Xuzhou (7.4%, n=7), and Yancheng (7.4%, n=7), with other cities having less than 5 strains in toto.
Antimicrobial resistance of S. sonnei
Our research validated an increasing resistance of S. sonnei to cefotaxime, with its resistance rate of 31.6% in 2012, significantly increasing to 64.3% in 2015, two times over that of 2012. During the four years, there was a rising tendency of resistance of S. sonnei to ampicillin, nalidixic acid, tetracycline and cotrimoxazole, with the mean rates of resistance to each agent all exceeding 70%. A significant decline in resistance to gentamicin was observed, with 31.6% in 2012 and 10.7% in 2015. Moreover, S. sonnei isolates retained high susceptibility of 5.3% and 8.4% to amoxicillin/clavulanic acid and norfloxacin, respectively.
Cefotaxime-resistant S. sonnei exhibited a regional diversity, with Suzhou contributing to the highest resistance rate (77.8%), followed by Nanjing and Yancheng (75.0% and 57.1%, respectively) in contrast to the null drug resistance in Xuzhou, Lianyungang and Suqian (Table 1).
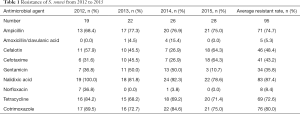
Full table
Molecular characterization
Of 95 clinical S. sonnei isolates, 66 strains were confirmed to have ESBLs-producing resistance genes by PCR amplification and DNA sequence analysis. 75 genotypes were detected in 66 S. sonnei isolates, with blaTEM-1 gene in 36 (48.0%) strains, and blaCTX-M-1 group in 28 (37.7%) strains, comprising 3 species of genotypes: blaCTX-M-55 (n=22), blaCTX-M-3 (n=3) and blaCTX-M-15 (n=3). There were 11 (14.7%) strains harboring blaCTX-M-9 group, encompassing blaCTX-M-14 (n=9) and blaCTX-M-65 (n=2). Both blaCTX-M-2 and blaSHV was negative in the amplification of these S. sonnei strains (Figure 1).
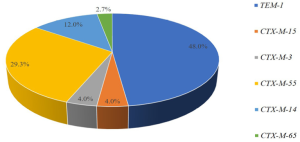
In addition, blaTEM-1 and blaCTX-M-55 were highly detectable in S. sonnei between 2012 and 2015, and the blaCTX-M-14 between 2013 and 2015, with the remainders less frequent (Table 2). In addition, with respect to cities in Jiangsu Province, blaTEM-1 (10 cities) was highly detectable, and blaCTX-M-55 (6 cities) and blaCTX-M-14 (5 cities) were moderate, whereas the remnants of genotypes were only detected in one city each.

Full table
Discussion
In China, shigellosis remains a common infectious disease, due to Shigella, a gram-negative, non-lactose-fermenting, and non-motile bacillus of the family Enterobacteriaceae. In the human body, Shigella specifically invades and colonizes the colonic mucosa, resulting in colonic disruption (4). As per the structure of the lipopolysaccharide O-antigen, Shigella is categorized into four species: S. flexneri, S. dysenteriae, S. boydii, and S. sonnei, with the most prevalent serotype being S. flexneri, followed by S. sonnei. Since the first collection and characterization by Sonne et al., S. sonnei was predominantly prevalent in developed countries, and global epidemiological studies showed a significant tendency in increasing prevalence, especially in developing countries. For instance, in China, the proportion of S. sonnei was 17.4% between 2003 and 2004, and reached 58.2% between 2011 and 2013, and was mainly distributed in the central and southeastern regions of China compared with S. flexneri (14). Therefore, our study was designed to investigate the prevalence and resistance pattern of S. sonnei in Jiangsu Province, China between 2012 and 2015 as well as the characterization of its molecular mechanisms, which might new light to the prophylaxis and clinical therapeutics for shigellosis.
A 10-year surveillance of antimicrobial susceptibility patterns among Shigella species isolated in China showed that Shigella strains resistant to cefotaxime increased from 7.87% in 2005 to 29.94% in 2014 (15). The resistance of S. sonnei to third-generation cephalosporin, particularly cefotaxime, significantly increased, has piqued our concern over resistance. Our data revealed a median resistance rate (43.2%) to cefotaxime in the S. sonnei from 2012 to 2015, well beyond that of S. sonnei (17.9%) monitored in Jiangsu Province between 2006 and 2011 (16). In addition, our findings also authenticated a rising tendency in resistance pattern, from 31.6% in 2012 to 64.3% in 2015, at a rate more than twice as that in 2012. Furthermore, in terms of provincial scales, the drug resistance rate of S. sonnei to cefotaxime was 30.8% in Zhejiang (17), 62.5% in Anhui (18), and 100% in Shanghai (19), far exceeding that in Jiangsu. Internationally, in Esfahān, Iran, S. sonnei showed a very high resistance pattern to cefotaxime, reaching 47.1% from 2010 to 2015 (20), with results similar to Jiangsu, China. In contrast, Nepal and Bangladesh, resistance rates to S. sonnei were relatively lower, accounting only 18.0% and 0%, respectively (21,22). Therefore, the continuous emergence of S. sonnei in Jiangsu Province and other developed provinces as well as the persistent elevation in drug resistance to cefotaxime might be attributed to the rapidity in economic development and the resultant modification of living styles and food diversity, wherefrom shigellosis cases inevitably occur and wherefore medication increased in these areas.
β-lactamase is the most indicated antimicrobial agent for Shigella, and plasmid-borne ESBLs, with the enzyme active against the expanded spectrum β-lactam antibiotics, consisting of four families, i.e., TEM, SHV, CTX-M and OXA and were produced by several members of the Enterobacteriaceae family. Our experimental data revealed that ESBL-encoding genes were mainly concentrated on the blaTEM-1, blaCTX-M-1 and blaCTX-M-9 group, whereas neither the blaCTX-M-2 or blaSHV-type was detectable. Moreover, blaTEM-1 (n=36, 48.0%) and blaCTX-M-55 (n=22, 29.3%) accounted for the largest proportion of ESBLs genotypes, indicating that the two subtypes were predominant types in Jiangsu Province, whereas the main epidemic ESBL-encoding genes were blaCTX-M-14 and blaCTX-M-15 in Zhejiang and Shanghai (17,19), and blaTEM-1 and blaCTX-M-14 in Anhui (23). In countries like Iran (24,25), Lebanon (26) and Turkey (27), blaCTX-M-15 has been the most prevalent ESBLs variant, thus posing a threat to human health (28).
blaTEM-1, described in 1965, was the first plasmid-mediated and the most commonly encountered β-lactamase in gram-negative bacteria. Amino acid substitutions of the TEM enzyme may possess the ability to hydrolyze cefotaxime, which may serve to explicate the production of TEM-1 β-lactamases in cefotaxime-resistant S. sonnei (29-31). Despite its slight proportion, TEM-1 β-lactamase in S. sonnei has been reported in several countries or regions, e.g., South Korea (32), Lebanon, Spain (33), Turkey as well as Anhui, China. In our study, 36 strains of S. sonnei harbor blaTEM-1, accounting for 37.9%, the largest proportion. By contrast to its modest proportions in other countries as well as other regions in China, blaTEM-1 detection rate increased at an alarming speed in Jiangsu, which requires vigilance against blaTEM-1 and countermeasures to avoid its sustained increase.
blaCTX-M-55, which is similar to blaCTX-M-15 and possesses only a single amino acid substitution, Ala-77-Val, was initially reported in ESBL-producing E. coli or K. pneumonia in Thailand (34). Subsequently, the blaCTX-M-55 gene was also detected in other gram-negative bacteria, such as Salmonella and Shigella (35,36). CTX-M-55 ESBLs are categorized under CTX-M-type β-lactamase, one of the most common β-lactamase-resistant ESBLs in China (37), for which CTX-M-55-type β-lactamase has been proposed to have hydrolytic activity and an increasing catalytic efficiency to cefotaxime (38). Our results show that of the 22 strains of S. sonnei harboring the blaCTX-M-55 gene, 21 strains presented resistance to cefotaxime, which was in conformance with the pre-conjecture mentioned afore. Not only blaCTX-M-55 constituted a large proportion in detection rate, but also exhibited a wide range of regional distribution, with its emergence in six cities in Jiangsu, validating its rapidity in dissemination in Jiangsu Province and awaiting meticulous surveillance.
Similar to the genes blaTEM-1 and blaCTX-M-55, CTX-M-14, is also a common ESBL subtype in Jiangsu Province, whereas other genotypes of ESBLs: blaCTX-M-3, blaCTX-M-15, and blaCTX-M-65, were detected in 3, 3 and 2 strains of S. sonnei, with each genotype detected in individual cities in this experiment. Despite their low detection rates, surveillance of these subtypes should be emphasized as well so as to prevent the dissemination of the resistance-producing genes between cities. In particular, two ESBL subtypes were coexistent in 10 strains of S. sonnei: 7 isolates were carriers of both blaTEM-1 and blaCTX-M-1 group genes, 2 isolates carried both blaTEM-1 and blaCTX-M-1 group genes, 1 strain harbored both blaCTX-M-1 and blaCTX-M-9 group genes, all of which affirmed the complexity and diversity of ESBLs-producing resistant genes of S. sonnei at the gene level.
Conclusions
Due to the increasing proportion of S. sonnei, in parallel with the rising resistance rate to cefotaxime, blaTEM-1 and blaCTX-M-55 of S. sonnei has become regional prevalent genotypes of ESBLs in Jiangsu Province. It is high time for each local healthcare and disease-control authority to implement surveillance of cefotaxime-resistant S. sonnei and bla genes and reduce the abuse of antibiotics with urgency, velocity and accuracy.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (81471994, 81702103), Jiangsu Provincial Natural Science Foundation (BK20151154, BK20170252), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six talent peaks project of Jiangsu Province (WSN-135), and Advanced health talent of six-one project of Jiangsu Province (LGY2016042).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Control and Prevention (IRB number: 2017018).
References
- Niyogi SK. Shigellosis. J Microbiol 2005;43:133-43. [PubMed]
- Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 1999;77:651-66. [PubMed]
- Ram PK, Crump JA, Gupta SK, et al. Analysis of data gaps pertaining to Shigella infections in low and medium human development index countries, 1984-2005. Epidemiol Infect 2008;136:577-603. [Crossref] [PubMed]
- Anderson M, Sansonetti PJ, Marteyn BS. Shigella Diversity and Changing Landscape: Insights for the Twenty-First Century. Front Cell Infect Microbiol 2016;6:45. [Crossref] [PubMed]
- Thompson CN, Duy PT, Baker S. The Rising Dominance of Shigella sonnei: An Intercontinental Shift in the Etiology of Bacillary Dysentery. PLoS Negl Trop Dis 2015;9:e0003708. [Crossref] [PubMed]
- Young LS. Review and Reassessment of dosing schedules for cefotaxime in selected medical indications. Diagn Microbiol Infect Dis 1995;22:147-54. [Crossref] [PubMed]
- Fullá N, Prado V, Durán C, et al. Surveillance for antimicrobial resistance profiles among Shigella species isolated from a semirural community in the northern administrative area of Santiago, Chile. Am J Trop Med Hyg 2005;72:851-4. [PubMed]
- Hosseini Nave H, Mansouri S, Sadeghi A, et al. Molecular diagnosis and anti-microbial resistance patterns among Shigella spp. isolated from patients with diarrhea. Gastroenterol Hepatol Bed Bench 2016;9:205-10. [PubMed]
- Knothe H, Shah P, Krcmery V, et al. Transferable Resistance to Cefotaxime, Cefoxitin, Cefamandole and Cefuroxime in Clinical Isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983;11:315-7. [Crossref] [PubMed]
- Kim S, Kim J, Kang Y, et al. Occurrence of extended-spectrum beta-lactamases in members of the genus Shigella in the Republic of Korea. J Clin Microbiol 2004;42:5264-9. [Crossref] [PubMed]
- Nguyen NT, Ha V, Tran NV, et al. The sudden dominance of blaCTX-M harbouring plasmids in Shigella spp. Circulating in Southern Vietnam. PLoS Negl Trop Dis 2010;4:e702. [Crossref] [PubMed]
- Clinical and Laboratory Standards Institute. M100-S22L Performance standards for antimicrobial susceptibility testing. Available online: http://www.facm.ucl.ac.be/intranet/CLSI/CLSI-M100S22-susceptibility%20testing-2012-original.pdf
- Yu Y, Ji S, Chen Y, et al. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect 2007;54:53-7. [Crossref] [PubMed]
- Qiu S, Xu X, Yang C, et al. Shift in serotype distribution of Shigella species in China, 2003–2013. Clin Microbiol Infect 2015;21:252.e5-8. [Crossref] [PubMed]
- Chang Z, Zhang J, Ran L, et al. The changing epidemiology of bacillary dysentery and characteristics of antimicrobial resistance of Shigella isolated in China from 2004–2014. BMC Infect Dis 2016;16:685. [Crossref] [PubMed]
- Gu B, Xu T, Kang H, et al. A 10-year surveillance of antimicrobial susceptibility patterns in Shigella sonnei isolates circulating in Jiangsu Province, China. J Glob Antimicrob Resist 2017;10:29-34. [Crossref] [PubMed]
- Zhang R, Zhou HW, Cai JC, et al. Serotypes and extended-spectrum β-lactamase types of clinical isolates of Shigella spp. from the Zhejiang province of China. Diagn Microbiol Infect Dis 2011;69:98-104. [Crossref] [PubMed]
- Yang H, Chen G, Zhu Y, et al. Surveillance of Antimicrobial Susceptibility Patterns among Shigella Species Isolated in China during the 7-Year Period of 2005-2011. Ann Lab Med 2013;33:111-5. [Crossref] [PubMed]
- Li J, Li B, Ni Y, et al. Molecular characterization of the extended-spectrum beta-lactamase (ESBL)-producing Shigella spp. in Shanghai. Eur J Clin Microbiol Infect Dis 2015;34:447-51. [Crossref] [PubMed]
- Mostafavi N, Bighamian M, Mobasherizade S, et al. Resistance of Shigella strains to extended-spectrum cephalosporins in Isfahan province. Med J Islam Repub Iran 2016;30:428. [PubMed]
- Shakya G, Acharya J, Adhikari S, et al. Shigellosis in Nepal: 13 years review of nationwide surveillance. J Health Popul Nutr 2016;35:36. [Crossref] [PubMed]
- Ud-Din AI, Wahid SU, Latif HA, et al. Changing trends in the prevalence of Shigella species: emergence of multi-drug resistant Shigella sonnei biotype g in Bangladesh. PLoS One 2013;8:e82601. [Crossref] [PubMed]
- Xiong Z, Li T, Xu Y, et al. Detection of CTX-M-14 extended-spectrum b-lactamase in Shigella sonnei isolates from China. J Infect 2007;55:e125-8. [Crossref] [PubMed]
- Bialvaei AZ, Kafil HS, Asgharzadeh M, et al. CTX-M extended-spectrum -lactamase-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz J Microbiol 2016;47:706-11. [Crossref] [PubMed]
- Ranjbar R, Ghazi FM, Farshad S, et al. The occurrence of extended-spectrum β-lactamase producing Shigella spp. in Tehran, Iran. Iran J Microbiol 2013;5:108-12. [PubMed]
- Sabra AH, Araj GF, Kattar MM, et al. Molecular characterization of ESBL-producing Shigella sonnei isolates from patients with bacilliary dysentery in Lebanon. J Infect Dev Ctries 2009;3:300-5. [PubMed]
- Kacmaz B, Unaldi O, Sultan N, et al. Drug resistance profiles and clonality of sporadic Shigella sonnei isolates in Ankara, Turkey. Braz J Microbiol 2014;45:845-9. [Crossref] [PubMed]
- Mehrgan H, Rahbar M. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli in a tertiary care hospital in Tehran, Iran. Int J Antimicrob Agents 2008;31:147-51. [Crossref] [PubMed]
- Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 1965;208:239-41. [Crossref] [PubMed]
- Bradford PA. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev 2001;14:933-51. [Crossref] [PubMed]
- Paterson DL, Bonomo RA. Extended-Spectrum β-Lactamases: A Clinical Update. Clin Microbiol Rev 2005;18:657-86. [Crossref] [PubMed]
- Lee JC, Oh JY, Kim KS, et al. Antimicrobial resistance of Shigella sonnei in Korea during the last two decades. APMIS 2001;109:228-34. [Crossref] [PubMed]
- Seral C, Rojo-Bezares B, Garrido A, et al. Characterization of a CTX-M-15-producing Shigella sonnei in a Spanish patient who had not travelled abroad. Enferm Infecc Microbiol Clin 2012;30:469-71. [Crossref] [PubMed]
- Kiratisin P, Apisarnthanarak A, Saifon P, et al. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum β-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis 2007;58:349-55. [Crossref] [PubMed]
- Yu F, Chen Q, Yu X, et al. High prevalence of extended-spectrum beta lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS One 2011;6:e16801. [Crossref] [PubMed]
- Qu F, Ying Z, Zhang C, et al. Plasmid-encoding extended-spectrum β-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol 2014;9:1143-50. [Crossref] [PubMed]
- Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect 2008;14 Suppl 1:33-41. [Crossref] [PubMed]
- Lee W, Chung HS, Lee H, et al. CTX-M-55-Type Extended-Spectrum β-lactamase Producing Shigella sonnei Isolated from a Korean Patient Who Had Travelled to China. Ann Lab Med 2013;33:141-4. [Crossref] [PubMed]


