Colorectal cancer genomics and designing rational trials
Genomics of colorectal cancer (CRC)
The description of the adenoma-carcinoma sequence by Vogelstein and Fearon in 1990 first led to an understanding of CRC as a disease of successive genomic changes (1) and subsequent genomic studies have refined and expanded this understanding. Among the most important milestones, The Cancer Genome Atlas (TCGA) network performed a comprehensive molecular characterization of 224 resected colon and rectal tumors, demonstrating similar patterns of genomic alterations in these tumors, identifying recurrent genomic alterations, and characterizing tumors based on mutation frequency as ultramutated, hypermutated, and non-hypermutated (2) and the CRC Subtyping Consortium analyzed gene expression profiles to describe a consensus molecular subtype (CMS) classification consisting of four subgroups based on results from six independent transcriptomic-based subtyping systems (3). Despite the rapidly advancing understanding of genomic alterations in CRCs, until recently the clinical utility of molecular information in standard practice was limited to the use of KRAS as a negative predictive biomarker of response to EGFR (epidermal growth factor receptor) targeting antibodies. With decreasing costs and turnaround time, next generation multi-gene sequencing panels of tumor tissue and circulating free DNA (cfDNA) have entered the clinic and the use of these methods has been rising exponentially (4). In this review, we will discuss the increasing incorporation of genomics in the treatment of patients with metastatic colorectal cancer (mCRC) both in the context of standard practice and clinical research.
Standard genomic markers in current clinical practice
Our knowledge of the genomic landscape of human cancers has rapidly accelerated with technological advances in sequencing, from capillary-based sequencing technologies to the modern massively parallel sequencing of today (5). Next generation sequencing (NGS) assays are highly sensitive, can analyze a large panel of genes, and detect novel mutations, small insertions and deletions (indels), copy number alterations, and select gene fusions and rearrangements from small amounts of DNA (6). A detailed description of technical aspects of NGS is beyond the aims of this review, but its fundamental principle is the spatial separation of individual DNA molecules and simultaneous analyses of millions of individual molecules. As each nucleotide in the sequences of each of the DNA strands is individually analyzed, the data are recorded and compiled computationally. The compiled data enable the bioinformatic analysis of multiple genes from multiple samples (7).
RAS
EGFR targeting antibodies, the first molecular targeted therapy for CRC, brought genomic data to the clinical assessment of CRC patients. These drugs improve survival in metastatic disease, but response is seen in only about 10% of unselected CRC cases (8,9). EGFR expression, the logical marker for these agents, was found not to correlate with tumor response (10), and thus a search began for predictive markers to guide patient selection. The major breakthrough in this area was the identification of KRAS exon 2 mutations as predictors of lack of benefit (11,12). KRAS, a small GTP-binding protein, lies downstream of EGFR and acts as signal switch whose activation engages effectors that control proliferation, differentiation, and survival (13). Subsequently, activating KRAS hotspot mutations in exons 3 and 4 and in NRAS were found to also predict for lack of benefit from EGFR therapies, refining the population of patients for these agents (14). Clinical data suggest that the use of EGFR inhibitors in patients with RAS mutant tumors may be associated with harm and shorter survival (14) and it has been speculated that this may be due to the inhibition of wild-type RAS within these tumors (15). The prognostic value of RAS mutations is more controversial, but increasing data associate RAS activating mutations in CRC with shorter survival and increased risk of recurrence after resection of metastases (16-18).
BRAF
BRAF encodes a serine/threonine protein directly downstream from RAS in the canonical mitogen-activated protein kinase (MAPK) pathway and mutations in this gene occur in up to 12% of mCRC patients (19). The majority of these consist of a substitution of glutamic acid for valine at the V600 hotspot in exon 15. The resultant V600E BRAF mutant is constitutively activated and can signal independent of RAS activation (20). The clinical validation of this biomarker contrasts with that of RAS, as BRAF V600E was demonstrated early on to be a strong negative prognostic biomarker in mCRC and later was appreciated as a negative predictive marker for EGFR inhibitor treatment. BRAF V600E mutation is associated with shorter overall survival, estimated around 14 months (21,22). This survival interval is similar to what was seen before modern combination chemotherapies, suggesting limited activity of second line treatment in this subgroup. BRAF V600E is associated with the poor prognostic features of T4 primary tumors, poor tumor differentiation, and peritoneal carcinomatosis (23-25). Whether the presence of this mutation should alter first line treatment remains controversial, but molecular subgroup analysis of the TRIBE trial suggests that for fit patients the combination FOLFOXIRI-bevacizumab can achieve better overall survival compared to FOLFIRI-bevacizumab (16).
In contrast to the strong prognostic effect of BRAF V600E, the predictive effect of BRAF V600E on EGFR inhibitor response has been less clear, likely due to the poor prognosis of this subgroup overall and a more subtle effect compared to RAS mutation; EGFR inhibitor treatment does not appear to cause harm in BRAF V600E mCRC, presumably due to the low levels of activated wild-type RAS in these tumors where high extracellular signal-regulated kinase (ERK) activation feedback suppresses upstream signaling. Three systematic reviews have evaluated BRAF V600E as a predictive marker for response to anti-EGFR therapy and all have found no significant benefit for EGFR inhibition in BRAF V600E mCRC (26-28). SWOG 1406 which tested irinotecan-cetuximab combination as a control arm in the second or third line setting in BRAF V600E mCRC found a response rate of 4% and median progression-free survival of 2 months to this combination, further supporting limited activity of cetuximab in this population (29). The National Comprehensive Cancer Network (NCCN) guidelines recognize that BRAF V600E mutation makes response to panitumumab or cetuximab highly unlikely (30).
It has recently become appreciated that not all BRAF mutants act the same (31). Two groups of non-V600 BRAF mutants have been described (32). The first group acts as constitutively activated proteins that signal as dimers, independent of RAS, and the second group consists of hypoactive BRAF mutants that bind more tightly than wild-type BRAF to RAS and exhibit enhanced binding and activation of wild-type CRAF to amplify upstream signaling. Hypoactive BRAF mutants have been associated with improved survival and increased sensitivity to EGFR inhibitors (32-34).
Differences in genomics between right and left primary tumors
The different ontogenesis of right- and left-sided colorectal tumors suggests biological differences within CRC, but it was not until recently that this variable started to be analyzed thoroughly in the metastatic setting. The two major clinical implications of primary tumor laterality are that right-sided tumors have a worse prognosis (35,36) and are insensitive to anti-EGFR therapy (37-39). The difference in prognosis appears to be driven by a higher rate of mitogenic mutations in right-sided tumors, with an increased frequency of RAS, PI3K, and TGFβ pathway alterations. Among microsatellite stable mCRC, 80% of tumors with a right-sided primary harbor a hotspot RAS mutation or BRAF V600E compared to 46% of left-sided primary tumors. Left-sided tumors often have no discernable mitogenic genomic driver, and growth factor ligand expression is higher in left colon primaries (40). Specifically, left-sided tumors express higher levels of amphiregulin and epiregulin, which are associated with increased sensitivity to EGFR antibodies (41). Microsatellite instability-high CRC, while rare in metastatic disease, occurs much more commonly in the right colon. All mCRC cases should be evaluated for microsatellite instability as immune checkpoint inhibitors are associated with high response rates in this population (42,43).
Genomic markers for targeted therapies in clinical trials
BRAF V600E
BRAF V600E is the most prevalent potentially targetable alteration in mCRC. RAF inhibitors are FDA approved for the treatment of BRAF V600 mutant melanoma, non-small cell lung cancer (NSCLC), and Erdheim-Chester disease (44,45). While RAF inhibitors have a high response rate in these diseases, these drugs have limited efficacy in BRAF V600E CRC as single agents (46). It is thought that high levels of basal receptor tyrosine kinase (RTK) signaling, particularly EGFR, in CRC underlie rapid adaptive resistance because ERK inhibition releases these receptors from negative feedback suppression (47,48). Receptor reactivation causes a rebound in ERK phosphorylation both by reactivating ERK signaling and by recruiting RAS which forms RAF dimers, which are insensitive to current RAF inhibitors (as these drugs selectively inhibit RAF monomers). Based on this understanding, clinical trials have tested combination therapies of RAF and EGFR inhibitors with modest response rates of 10–25% (29,44,49-52) (Table 1). Adding a mitogen-activated protein kinase (MEK) inhibitor to RAF and EGFR inhibitors leads to more profound inhibition of ERK signaling and improved response rates (53). The ongoing phase 3 BEACON trial will evaluate whether doublet or triplet targeted therapy extends survival compared to standard therapy containing irinotecan and cetuximab for patients with BRAF V600E mCRC. Early results from the safety lead-in (n=29) for the triplet combination of the RAF inhibitor encorafenib, MEK inhibitor binimetinib, and cetuximab showed an overall response rate of 48% in patients with BRAF V600E mCRC (54).
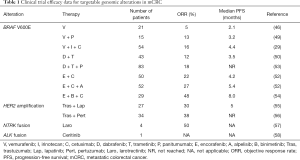
Full table
While combination therapy has increased response rates in mCRC, most patients progress within a few months of starting targeted therapy. Reactivation of ERK signaling, rather than alterations in parallel signaling pathways, appears to underlie resistance in patient tumors (59,60). ERK inhibitors and novel RAF inhibitors that inhibit mutant monomers and dimers have been proposed as new agents that may be able to overcome resistance (59,60).
HER2 amplification
Amplification of ERBB2 (also called HER2) occurs in 2% to 6% of mCRC and is associated with insensitivity to EGFR antibody treatment (61-63). Targeting this RTK with the anti-HER2 antibody trastuzumab is one of the major successes in the treatment of breast cancer and since then further anti-HER2 drugs have demonstrated benefit in this disease. Treatment of colon cancer xenografts with HER2 amplification indicated that trastuzumab and lapatinib, a small molecular inhibitor of EGFR/HER2 did not cause tumor regression as monotherapy, but the combination demonstrated tumor shrinkage (64). This combination was studied in the phase 2 HERACLES study in 27 patients with HER2-positive mCRC and 30% of patients achieved an objective response (55). Cohort B of the HERACLES trial, which is currently ongoing, will evaluate the activity of the antibody-drug conjugate TDM-1 and TDM-1 together with the anti-HER2 antibody pertuzumab in the second line setting. The multi-basket MyPathway study included a HER2 amplified mCRC cohort treated with trastuzumab plus pertuzumab of 37 patients and found a response rate of 38% (56). The clinical significance of HER2 mutations in mCRC has been less studied, and HER2 mutations are more commonly concurrent with RAS mutations in mCRC than is HER2 amplification. In two CRC patient-derived xenografts (PDXs) with HER2 mutations, treatment with trastuzumab plus the pan-HER inhibitor neratinib or lapatinib produced tumor regression (65). There is initial evidence of efficacy in HER2-mutated NSCLC and breast cancer patients treated with targeted therapy (66,67), but if these results will be replicated in mCRC remains to be seen.
NTRK fusions
The NTRK family of kinases includes three genes, NTRK1, NTRK2, and NTRK3, which encode three transmembrane receptors—TRKA, TRKB, and TRKC, respectively. Fusions involving the NTRK genes have been identified, at low frequencies, across various tumor types including CRC and involve numerous partners (68). Selective NTRK inhibitors, such as larotrectinib and entrectinib, have been associated with high response rates in tumors with these fusions (57), including responses seen in mCRC patients (69).
ALK, ROS, and RET fusions
Recurrent gene fusions involving ALK and ROS have been described primarily in NSCLC, but infrequently are present in other tumors types, including CRC (70,71). Targeted therapies against ALK or ROS kinase, including crizotinib, ceritinib, and alectinib, are FDA-approved for NSCLC with these alterations. In CRC, these fusions are present in 0.2% to 2.4% of patients (72,73). An analysis by Foundation Medicine of 3,117 advanced CRC cases genotyped with their NGS panel identified 6 cases (0.2%) with ALK fusions. One of these patients was treated with ceritinib and achieved a partial response lasting 9 months, until the tumor acquired a KRAS mutation (58).
RET kinase fusions are frequent in papillary thyroid cancer and have been described rarely also in lung adenocarcinoma and chronic myelomonocytic leukemia. In the Foundation Medicine series of 3,117 advanced CRC cases, RET fusions were present in 6 cases (0.2%). None of these patients had another driver mutation, and one of them was treated with regorafenib, which has activity against RET kinase. This patient had clinical and CEA tumor marker responses, but unfortunately no further follow-up was available as the patient died of unrelated causes shortly thereafter (74). In RET-rearranged lung adenocarcinoma drugs such as cabozantinib and lenvatinib have demonstrated initial activity (75,76), but in RET-rearranged mCRC these multikinase inhibitors have not been studied.
Designing rational trials
As noted above, currently targetable genomic alterations in mCRC are found in only a minority of cases and it has been difficult to bring targeted therapy for matched genomic alterations in mCRC to the clinic. Here we discuss some of the challenges and lessons learned in the delivery of precision medicine for mCRC.
Both genomic alteration and tissue histology influence response to targeted therapy
Where the same genomic alteration has been identified in mCRC and other tumor types, differing responses have been seen in mCRC compared to other tumor types. This is most dramatic for V600E BRAF where the BRAF inhibitors vemurafenib or dabrafenib are associated with a response rate of over 50% in melanoma and less than 10% in mCRC and combined RAF and MEK inhibitors, for example dabrafenib plus trametinib, achieves responses of about 70% in melanoma and 12% in mCRC (50,77). Also HER2 amplified mCRC appears relatively insensitive to single agent trastuzumab and dual HER2 inhibitor therapy is needed for responses, while trastuzumab alone is effective in treatment of HER2 amplified breast and gastric cancers (78,79). These differences have been attributed to the high RTK environment of the colorectum, where the activity of the targeted therapy is attenuated by release of negative feedback loops, reactivation of receptors, and a rebound in pathway signaling. Thus combination therapy is required to effectively inhibit pathway signaling and achieve clinical benefit. For example, in BRAF V600E mCRC, analysis of phosphorylated ERK expression levels before treatment exposure and after 14 days of treatment in paired tumor biopsies showed greater ERK inhibition with the triplet of RAF, MEK, and EGFR inhibition (dabrafenib, trametinib, and panitumumab) compared to baseline or to RAF and EGFR inhibition alone (dabrafenib and panitumumab), mirroring the improved response rate for the triplet, but the degree of ERK inhibition achieved with the triplet (60%) remained less than that seen for RAF inhibition (dabrafenib) in melanoma (84%) (53), providing a mechanistic explanation for the difference in activity between these tumor types. These data suggest that the activated oncogene (e.g., V600E BRAF or amplified HER2) remains a driver in CRC, but combination strategies that effectively inhibit the signaling network in mCRC must be devised to achieve greater clinical benefit.
Resistance is dynamic and often involves subclonal populations
The most common mechanism of acquired resistance to EGFR inhibitors in mCRC patients is the emergence of KRAS hotspot mutations (80-82). These mutants are thought to emerge from selection of a minor, pre-existing clone (83). Analysis of cfDNA in patients with progression through EGFR inhibitors indicates that the prevalence of these KRAS mutations in cfDNA is lower than that seen in patients with baseline KRAS mutant mCRC and the percent mutant reads decreases with time from last EGFR inhibitor exposure (82). These data suggest that resistance results from selection of subclonal populations and is dynamic based on exposure to the selective pressure. In patients with BRAF V600E mCRC progressing through targeted therapies, serial studies of cfDNA identified the emergence of more than one RAS mutation in 25% of patients (53), consistent with multiple, subclonal, resistant populations. In a patient at Memorial Sloan Kettering Cancer Center with BRAF V600E mCRC treated with vemurafenib/panitumumab who developed resistance with a newly detected NRAS Q61K mutation (60), the resistance alteration became undetectable off treatment, but reemerged after 8 weeks of rechallenge with vemurafenib and panitumumab (Figure 1). These data suggest clinical trials with intermittent schedules may be able to forestall resistance (84).
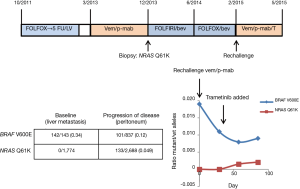
Adaptive trial design with real-time monitoring of tumor subpopulations
The initial experience with targeted therapy in mCRC recommends a future of drug development with real-time pharmacodynamic markers and with tracking of sensitive and resistant tumor populations, for example with circulating tumor DNA (ctDNA) analysis. The best treatment regimens will need to profoundly inhibit the target while minimizing toxicity, perhaps by taking advantage of real-time cfDNA analysis guiding intermittent dosing schedules. As RTK signaling plays a key role in relative resistance in the colorectum, future adaptive designs may further refine patient subpopulations by testing personalized combination regimens aimed at inhibiting the activated oncogene and the dominant reactivated receptor.
Conclusions
CRC results from successive genomic and epigenetic alterations, but it has not been straightforward to realize the promise of targeted therapies in mCRC. Often it has not been sufficient to directly inhibit the target, but rather addressing the signaling network that feeds into the target is required. At this time, negative predictive markers for response to EGFR targeted therapies and the positive predictive marker of microsatellite instability for response to immune checkpoint blockade are part of standard care. Emerging data support a developing role for targeted therapies in mCRC with HER2 amplification, NTRK rearrangements, or BRAF V600E mutations. Real-time pharmacodynamic and cfDNA analysis will allow further refining and optimizing targeted therapy approaches for mCRC.
Acknowledgements
Funding: Supported by the National Institutes of Health Memorial Sloan Kettering Cancer Center Core Grant (P30 CA 008748).
Footnote
Conflicts of Interest: R Yaeger has served on an advisory board for GlaxoSmithKline. S Mondaca has no conflicts of interest to declare.
References
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Hyman DM, Taylor BS, Baselga J. Implementing Genome-Driven Oncology. Cell 2017;168:584-99. [Crossref] [PubMed]
- Atreya CE, Yaeger R, Chu E. Systemic Therapy for Metastatic Colorectal Cancer: From Current Standards to Future Molecular Targeted Approaches. Am Soc Clin Oncol Educ Book 2017;37:246-56. [Crossref] [PubMed]
- Sheikine Y, Kuo FC, Lindeman NI. Clinical and Technical Aspects of Genomic Diagnostics for Precision Oncology. J Clin Oncol 2017;35:929-33. [Crossref] [PubMed]
- Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040-8. [Crossref] [PubMed]
- Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658-64. [Crossref] [PubMed]
- Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803-10. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 1990;348:125-32. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Berlin J. Beyond exon 2--the developing story of RAS mutations in colorectal cancer. N Engl J Med 2013;369:1059-60. [Crossref] [PubMed]
- Cremolini C, Loupakis F, Antoniotti C, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol 2015;16:1306-15. [Crossref] [PubMed]
- Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013;258:619-26; discussion 626-7. [Crossref] [PubMed]
- Cercek A, Braghiroli MI, Chou JF, et al. Clinical Features and Outcomes of Patients with Colorectal Cancers Harboring NRAS Mutations. Clin Cancer Res 2017;23:4753-60. [Crossref] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010;464:427-30. [Crossref] [PubMed]
- Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-9. [Crossref] [PubMed]
- Saridaki Z, Papadatos-Pastos D, Tzardi M, et al. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients' outcome. Br J Cancer 2010;102:1762-8. [Crossref] [PubMed]
- Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011;117:4623-32. [Crossref] [PubMed]
- Clancy C, Burke JP, Kalady MF, et al. BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: a systematic review and meta-analysis. Colorectal Dis 2013;15:e711-8. [Crossref] [PubMed]
- Yaeger R, Cercek A, Chou JF, et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 2014;120:2316-24. [Crossref] [PubMed]
- Rowland A, Dias MM, Wiese MD, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer 2015;112:1888-94. [Crossref] [PubMed]
- Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51:587-94. [Crossref] [PubMed]
- van Brummelen EM, de Boer A, Beijnen JH, et al. BRAF Mutations as Predictive Biomarker for Response to Anti-EGFR Monoclonal Antibodies. Oncologist 2017;22:864-72. [Crossref] [PubMed]
- Kopetz S, McDonough SL, Morris VK, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG 1406). J Clin Oncol 2017;35:abstr 520.
- National Comprehensive Cancer Network Colon Cancer Version 2.2017. Accessed November 20, 2017. Available online: www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- Jones JC, Renfro LA, Al-Shamsi HO, et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol 2017;35:2624-30. [Crossref] [PubMed]
- Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234-8. [PubMed]
- Summers MG, Smith CG, Maughan TS, et al. BRAF and NRAS Locus-Specific Variants Have Different Outcomes on Survival to Colorectal Cancer. Clin Cancer Res 2017;23:2742-9. [Crossref] [PubMed]
- Cremolini C, Di Bartolomeo M, Amatu A, et al. BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis. Ann Oncol 2015;26:2092-7. [Crossref] [PubMed]
- Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 2015;107:107. [Crossref] [PubMed]
- Stintzing S, Tejpar S, Gibbs P, et al. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer 2017;84:69-80. [Crossref] [PubMed]
- Brulé SY, Jonker DJ, Karapetis CS, et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer 2015;51:1405-14. [Crossref] [PubMed]
- Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA Oncol 2016. [Epub ahead of print]. [PubMed]
- Venook AP, Niedzwiecki D, Innocenti F, et al. Impact of primary (1º) tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol 2016;34:abstr 3504.
- Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33:125-36.e3. [Crossref] [PubMed]
- Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007;25:3230-7. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182-91. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [Crossref] [PubMed]
- Kopetz S, Desai J, Chan E, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol 2015;33:4032-8. [Crossref] [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [Crossref] [PubMed]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227-35. [Crossref] [PubMed]
- Yaeger R, Cercek A, O'Reilly EM, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin Cancer Res 2015;21:1313-20. [Crossref] [PubMed]
- Corcoran RB, Atreya CE, Falchook GS, et al. Combined BRAF and MEK Inhibition With Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J Clin Oncol 2015;33:4023-31. [Crossref] [PubMed]
- van Geel RM, Tabernero J, Elez E, et al. A Phase Ib Dose-Escalation Study of Encorafenib and Cetuximab with or without Alpelisib in Metastatic BRAF-Mutant Colorectal Cancer. Cancer Discov 2017;7:610-9. [Crossref] [PubMed]
- Tabernero J, Geel RV, Guren TK, et al. Phase 2 results: Encorafenib (ENCO) and cetuximab (CETUX) with or without alpelisib (ALP) in patients with advanced BRAF-mutant colorectal cancer (BRAFm CRC). J Clin Oncol 2016;34:abstr 3544.
- Corcoran RB, André T, Atreya CE, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF V600E-mutant colorectal cancer. Cancer Discov 2018;8:428-43. [Crossref] [PubMed]
- Cutsem EV, Cuyle PJ, Huijberts S, et al. BEACON CRC study safety lead-in (SLI) in patients with BRAFV600E metastatic colorectal cancer (mCRC): Efficacy and tumor markers. J Clin Oncol 2018;36:abstr 627.
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:738-46. [Crossref] [PubMed]
- Hainsworth JD, Meric-Bernstam F, Swanton C, et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from MyPathway, an Open-Label, Phase IIa multiple basket study. J Clin Oncol 2018;36:536-42. [Crossref] [PubMed]
- Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. [Crossref] [PubMed]
- Yakirevich E, Resnick MB, Mangray S, et al. Oncogenic ALK Fusion in Rare and Aggressive Subtype of Colorectal Adenocarcinoma as a Potential Therapeutic Target. Clin Cancer Res 2016;22:3831-40. [Crossref] [PubMed]
- Ahronian LG, Sennott EM, Van Allen EM, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 2015;5:358-67. [Crossref] [PubMed]
- Yaeger R, Yao Z, Hyman DM, et al. Mechanisms of acquired resistance to BRAF V600E inhibition in colon cancers converge on RAF dimerization and are sensitive to its inhibition. Cancer Res 2017;77:6513-23. [Crossref] [PubMed]
- Nathanson DR, Culliford AT, Shia J, et al. HER 2/neu expression and gene amplification in colon cancer. Int J Cancer 2003;105:796-802. [Crossref] [PubMed]
- Seo AN, Kwak Y, Kim DW, et al. HER2 status in colorectal cancer: its clinical significance and the relationship between HER2 gene amplification and expression. PLoS One 2014;9:e98528. [Crossref] [PubMed]
- Martin V, Landi L, Molinari F, et al. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668-75. [Crossref] [PubMed]
- Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508-23. [Crossref] [PubMed]
- Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015;5:832-41. [Crossref] [PubMed]
- Ben-Baruch NE, Bose R, Kavuri SM, et al. HER2-Mutated Breast Cancer Responds to Treatment With Single-Agent Neratinib, a Second-Generation HER2/EGFR Tyrosine Kinase Inhibitor. J Natl Compr Canc Netw 2015;13:1061-4. [Crossref] [PubMed]
- Li BT, Shen R, Buonocore D, et al. Ado-trastuzumab emtansine in patients with HER2 mutant lung cancers: Results from a phase II basket trial. J Clin Oncol 2017;35:abstr 8510.
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Sartore-Bianchi A, Ardini E, Bosotti R, et al. Sensitivity to Entrectinib Associated With a Novel LMNA-NTRK1 Gene Fusion in Metastatic Colorectal Cancer. J Natl Cancer Inst 2015;108. [Crossref] [PubMed]
- Shaw AT, Hsu PP, Awad MM, et al. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013;13:772-87. [Crossref] [PubMed]
- Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 2014;5:3116. [Crossref] [PubMed]
- Rankin A, Klempner SJ, Erlich R, et al. Broad Detection of Alterations Predicted to Confer Lack of Benefit From EGFR Antibodies or Sensitivity to Targeted Therapy in Advanced Colorectal Cancer. Oncologist 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Aisner DL, Nguyen TT, Paskulin DD, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 2014;12:111-8. [Crossref] [PubMed]
- Le Rolle AF, Klempner SJ, Garrett CR, et al. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget 2015;6:28929-37. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]
- Long GV, Stroyakovskiy D, Gogas H, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 2015;386:444-51. [Crossref] [PubMed]
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783-92. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532-6. [Crossref] [PubMed]
- Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 2016;7:13665. [Crossref] [PubMed]
- Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26:731-6. [Crossref] [PubMed]
- Diaz LA, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [Crossref] [PubMed]
- Das Thakur M, Salangsang F, Landman AS, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 2013;494:251-5. [Crossref] [PubMed]


