MicroRNAs as biomarkers of acute lung injury
Introduction
The acute respiratory distress syndrome (ARDS), as defined by the Berlin consensus conference, is a type of acute diffuse lung injury associated with a predisposing risk factor, characterized by inflammation leading to increased pulmonary vascular permeability and loss of aerated lung tissue. Clinical (hypoxemia, bilateral radiographic opacities, and physiological derangements such as increased pulmonary venous admixture, increased physiological dead space and decreased respiratory system compliance) and morphological hallmarks [changes consistent with diffuse alveolar damage (DAD)] characterize the syndrome (1). However, the clinical criteria for the diagnosis of ARDS as defined in the Berlin definition (2) are poor for the identification of the clinic-pathological entity (i.e., diffuse lung injury and DAD) (3-7).
Current evidence indicates that the identification of DAD in the group of patients fulfilling the clinical criteria for the diagnosis of ARDS is of clinical relevance, as the clinical course of patients with ARDS differs in those with DAD and in those without DAD (i.e., higher mortality and higher degree of organ dysfunction in patients DAD versus patients without DAD) (7-10). On the other hand, enrichment of the treatment group with patients with a homogeneous phenotype will increase the probability of finding positive results in clinical trials on the therapeutic effects of certain interventions (11).
Biomarkers of ARDS
Biomarkers can be used for identification of homogeneous phenotypes within a given condition, risk stratification, prognostication of specific outcomes (e.g., mortality), assessment of the degree of disease activity, prediction of response to therapy, or monitoring response to therapy.
Using biomarkers for the diagnosis of ARDS or for the identification of patients that are more likely to benefit from certain therapeutic interventions will have a major impact in clinical practice and in the development of new diagnostic and therapeutic tools (12,13). Numerous studies have analyzed the role of different biomarkers of ARDS, but none has been proven to be reliable enough to be used in clinical practice for one of the possible uses mentioned above (14-17).
Studies have identified patients with the diagnosis of ARDS based on clinical criteria, despite the fact that different phenotypes are included under these criteria. Thus, a translational approach is needed for the design of studies in which the aim is the identification of the clinicopathological entity, including its pathological manifestation (i.e., DAD), with the difficulty that in most cases obtaining lung tissue for histological confirmation is not feasible.
In the present review, we discuss the role of microRNAs (miRNAs) as biomarkers of ARDS and DAD, and their potential use as therapeutic targets for this condition.
miRNAs biogenesis and function
miRNAs are a class of small (19–24 nucleotides in length), non-coding single-stranded RNAs with a fundamental role in post-transcriptional regulation of gene expression. They target the 3’untranslated regions (UTRs) of >60% for human genes, promoting their degradation or attenuating their translation (18,19). iRNAs exert their action through regulatory mechanisms post-transcriptionally. They bind the complementary 3’untraslated or open reading frame regions (ORFR) of target messenger RNAs (mRNAs) generating its degradation or the downregulation of protein expression (20). miRNAs can be produced from their own genes (canonical or non-canonical pathways) or from introns of other genes (miRtron). The fact that most miRNA genes are in intergenic regions or oriented antisense to neighboring genes supports the hypothesis that their transcription can be specifically regulated (independent transcriptional units). In other cases, the miRNA gene is transcribed together with its host gene and the expression regulation is the same for both genes (miRNA and host gene). miRNA genes are usually transcribed by RNA polymerase II (polymerase III in the case of some miRNAs) and then processed into mature miRNAs through canonical or non-canonical miRNA biogenesis pathways (Figure 1). Such transcripts may be monocistronic (with only one hairpin) or polycistronic (several hairpins). The presence of multiple genes in the same transcript (polycistronic transcripts) is very common in prokaryote cells but extremely uncommon in eukaryotes. This special feature allows the simultaneous regulation of a cluster of genes.
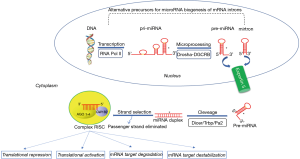
First, a primitive miRNA (100–1,000 nucleotides, called pri-miRNA) is generated and further processed to form shorter precursors (about 70 nucleotides, now called pre-miRNA) by an RNase type III enzyme called “Drosha” along with at least 20 other polypeptides, such as the critical region protein Gene 8 of the DiGeorge syndrome (Pasha in plants) (21). Alternatively, some noncanonical biogenesis pathways may occur during mRNA splicing giving rise to “miRtrons”. miRtrons are, in fact, the spliced-out introns of mRNAs, which constitute functional pre-miRNAs (22).
Subsequently to the formation of pre-miRNA, these are transported from the nucleus to the cytoplasm, which is facilitated by the exportin-5 protein. In the cytoplasm, the pre-miRNA is cleaved by Dicer (RNase III), generating a miRNA duplex of approximately 22 nucleotides containing the mature miRNA and the so-called miRNA* (23). These miRNA: miRNA* duplex gives rise to two types of sequences: the 5p sequence and the complementary 3p sequence (designated in the old nomenclature with an asterisk). Although 3p miRNAs were initially considered as a waste product, they have been recently shown to play significant roles in cellular function and to be associated with human disease (24).
The miRNA: miRNA* duplex is loaded in a multiprotein complex called RNA-induced silencing complex (RISC). In the canonical processing of miRNA, one of the chains is taken by the Argonaute proteins (Ago) which are key catalytic enzymes that facilitate incorporation of the mature miRNA into an RISC. The RISC complex is comprised of several proteins and mature miRNA strand.
Post-transcriptional regulation is achieved predominantly by miRNA-RISC complex binding in the 3’UTR of the target mRNA. The function of the mature miRNA strand is to recognize by complementarity the mRNA which will be broken and degraded by the action of Ago. This process is called RNA interference and is considered one of the most potent post-transcriptional mechanisms regulating gene expression (25). The complementarity of the bases between the miRNA and the mRNA determines the result of this repression: whereas a complete pairing produces a degradation of the target mRNA, an imperfect pairing leads to a sequestration of the target mRNA by the “P bodies” within the cytoplasm (26,27). It should be noted that there is a “seed” sequence that must be strictly complementary to the target mRNA (2–8 nucleotides at the 5’ end of a mature miRNA) (28). “Seed” sequences are conserved in many species and there is variation depending on the genes they regulate, and they are generally used to classify miRNA families. Mutations or changes in the length of the seed sequence in the seed region may show dramatic changes in gene expression, leading to pathological alterations such as cancer or coronary artery disease (29-32).
The miRNAs represent a novel pathway for post-regulatory regulation of gene expression, by blocking translation or inducing degradation of target mRNAs. This process has been proposed to be the major mechanism controlling tissue- and cell type-specific expression of miRNAs (19). Repression or degradation of the target mRNA prevents protein production, impacting on biological processes as important as proliferation, differentiation, apoptosis, etc. miRNAs may exert control on hundreds of mRNAs as they may only show partial complementarity with most target mRNAs to be able to modulate inhibe translation.
miRNAs as biomarkers
miRNAs are good candidates as disease biomarkers: (I) their expression changes in various disease states such autoimmune, neurodegenerative and inflammatory diseases (33,34); (II) they can be isolated from a wide range of body fluids; (III) they resist extreme environmental conditions; (IV) they participate in the early stages of gene expression, so that changes in miRNAs expression precede their subsequent effects; (V) they can be measured using readily available technology (23); (VI) they are potential therapeutic targets as each miRNA regulates the expression of many genes; (VII) their interference is feasible (35).
There are numerous evidences of alterations in the expression patterns (at the cellular and tissue level and in biological fluids) of miRNAs associated with key pathological processes in many diseases such as cardiovascular diseases, neural diseases, diabetes, cancer and inflammatory conditions (36-46).
Four additional characteristics make miRNAs excellent candidates as disease biomarkers. First, the expression profiles of miRNAs in different conditions appear to be unique and do not depend on age, race or sex (47). Second, the sequences of most miRNAs are conserved among different species. Third, the expression of some miRNAs appears to be highly specific to tissue or biological state. For example, miR-122 is preferentially expressed in liver and miR-133 in muscle (48,49). Fourth, the experimental manipulation of different miRNAs that are associated with malignancies modulates the malignant characteristics of tumor cell lines as well as the development of tumors in in vivo mouse models (50-53). Fifth, different individual studies confirm that circulating levels of miRNAs regain basal levels following the corresponding therapeutic treatment, demonstrating their potential utility as biomarkers of efficacy of a treatment (49).
In this context, we can hypothesize in the field of ARDS that the identification of miRNAs associated with this syndrome, not only would facilitate our understanding of the physiological mechanisms involved, but also could help identify potential therapeutic targets.
Circulating miRNAs as biomarkers
In 2008, it was discovered that miRNAs are not only located within cells but also in different biological fluids such as blood, serum, plasma, urine and bronchoalveolar fluid (43). It is well known that these extracellular miRNAs are very stable to the action of the ribonucleases that are present in serum or plasma (54). This protection, which makes miRNAs so stable, is due to the fact that circulating miRNAs are packaged in different extracellular vesicles (EVs), of which there are three types, according to their size and biogenesis: exosomes, microvesicles, and apoptotic bodies (Figure 2) (55-57). Exosomes originate from the endosomal pathway and form intracellular multivesicular bodies that fuse to the plasma membrane and are secreted as vesicles measuring 0.05–0.16 µm. Microvesicles arise through budding of the plasma membrane measuring 0.2–1 µm (58,59). Apoptotic bodies are released from blebbing of the plasma membrane of apoptotic cells and have diameters of 0.5–2 µm (60). They can also be transported by complexing with lipoproteins, mostly high-density lipoprotein (HDL) or associated with binding proteins such as Ago2 or nucleoplasmin 1 (NMP1), which protect them from possible degradation (61).
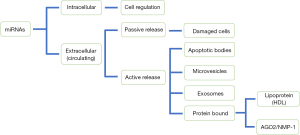
The presence of miRNAs in microparticles has generated the idea that these circulating miRNAs may be playing important roles in cellular communication, acting in autocrine, paracrine and endocrine communication systems regulating gene expression and the phenotype of the recipient cells (62). miRNAs must be packaged on the appropriate conveyors and actively secreted in a very selective manner. In addition, they can be released passively by damaged or necrotic cells (Figure 2) (63,64). Also, miRNAs must be transferred to specific receptor cells and more importantly, these miRNAs must retain the ability to recognize and repress their target mRNAs within the recipient cells (65-70).
miRNAs and the pathogenesis of ARDS
Almost every biological process, including development, hemostasis and inflammation, is regulated by miRNAs. Most of the studies on the role of miRNAs in pulmonary disease and development are based in research in animal models, and the degree of translation into human medicine is unclear. Some studies have investigated the potential role of miRNAs in the pathogenesis of acute lung injury (ALI)/ARDS. These studies show that certain miRNAs are significantly up-regulated or down-regulated in this condition or in related processes such as inflammation or apoptosis (Table 1). Modifications in the expression of different miRNAs regulate the inflammatory response and tissue repair mechanisms in ALI/ARDS, as these processes are concomitant to increased levels of inflammatory mediators, such as the pro-inflammatory cytokines IL-1β, TNF-α, IL-6 and IL-8, and the anti-inflammatory molecules IL-1, IL-10 and IL-13 (71-73).

Full table
Cai et al. studied for the first time the role that miRNAs could play in ALI. In a mouse model of lipopolysaccharide (LPS)-induced lung injury they found that mir-214 and miR-451 were up-regulated whereas mir-16, miR-23a, miR-24, miR-181a, miR-181b, and miR-199a were significantly down-regulated (74). On the other hand, they found in LPS-stimulated A549 lung cells that miR-16 regulates transcription of IL-6 and TNF-α (74).
Recently, Li et al. found that down-regulation of miR-181a protects mice from LPS-induced ALI. They suggest that miR-181a inhibition protects mice from LPS-induced lung injury by targeting Bcl-2, a key molecule in the regulation of cellular apoptosis (75). miRNA-1246 appears to mediate apoptosis of LPS-induced pulmonary endothelial cells through the regulation of the angiotensin-converting enzyme 2 (ACE-2), a potential biomarker of ARDS (76). Depletion of miR-1246 attenuated lung inflammation, neutrophil infiltration, and vascular permeability and restored pulmonary expression of ACE-2 in LPS-exposed mice. In another study, Xie et al. showed in an animal model of ALI that miR-127 significantly reduced lung permeability, infiltration of inflammatory cells, and proinflammatory cytokine levels (77). In addition, miR-127 is capable of modulating the polarization of macrophages and promoting pulmonary inflammation by stimulating proinflammatory cytokines (78). Recently, it has been suggested that upregulation of miR-127 expression may contribute to the development of ventilator-induced lung injury (VILI). This process would be mediated by an increase in the inflammatory response involving NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) and P38 mitogen-activated protein kinases-associated signaling pathways (79).
Vaporidi et al. studied pulmonary miRNA profiling in a mouse model of VILI (80). They reported an increase in the expression of 50 miRNAs and a decrease in the expression of 15 miRNAs from a total of 335 miRNAs detected. Inhibition of miR-21 avoided the deterioration in lung compliance (80).
Cheng et al. reported that LPS stimulation of mouse Raw 264.7 macrophage cells resulted in the up-regulation miR-146a and miR-155 and down-regulation of miR-27a-5p and miR-532-5p (81). In a similar cell model, Tili et al. showed up-regulation of miR-155 and down-regulation of miR-125b (82).
The relevance of miRNAs in ALI has been shown in sepsis-induced ALI. miR-133 is one of the most intensely studied and best characterized to date. This miRNA appears to be involved in processes such as fibrosis and inflammation (83,84). Concentrations of circulating miR-133 were significantly elevated in a large cohort of critically ill patients with sepsis versus healthy controls. This high level indicated poor survival and were predictor of mortality, making it a promising biomarker for the diagnosis and follow-up of sepsis-induced ALI (84). miR-27a also appears to play an important role in regulating the inflammatory response induced by sepsis as well as a promising target in the treatment and investigation of sepsis-induced ALI/ARDS. Wang et al. showed in an animal model of sepsis induced by cecal ligation and puncture that the inhibition of miR-27a expression significantly down-regulated the expression of TNF-α and IL-6 by reducing the phosphorylation level of NF-κB p65 subunit and blocking its function, hindering the ability to bind DNA. On the other hand, levels of peroxisome proliferator-activated receptor ɣ (PPARɣ), which can inhibit production of proinflammatory cytokines, were also positively regulated (85).
miR-150 is one of the first miRNAs that were studied in patients with critical illness and sepsis. This miRNA is preferably expressed in cells of the immune system, and thus could be a marker of the immune cell activation that occurs during inflammation and sepsis (86). Levels of expression in cell lines such as leukocytes stimulated with LPS showed down-regulation of miR-150 (87). Vasilescu et al. found lower levels of miR-150 in a cohort of 16 patients with abdominal sepsis and these low levels of miR-150 were correlated to increased levels of pro-inflammatory cytokines (88). More recently, the expression of miR-150 was analyzed in a larger cohort of 223 critically ill patients vs. 76 healthy controls. Lower concentrations of miR-150 were found in patients with sepsis, although a significant difference with healthy controls was not achieved (89).
miR-155 and miR-146a have important roles in inflammation. Wang et al. identified miR-155 as a pro-inflammatory factor as miR-155 gene inactivation protected mice from LPS-induced ALI (90). On the other hand, in vitro experiments performed in isolated alveolar macrophages confirmed that miR-155 expression was increased in response to LPS stimulation (90). Also, miR-155 antisense oligonucleotides resulted in a significant decrease in pro-inflammatory cytokines, such as TNF-α, IL-12 and monocyte chemotactic peptide-1 (MCP-1) in T-cells, whereas IL-10, an anti-inflammatory cytokine, notably increased (91).
miR-146a regulates Toll-like receptors (TLRs) and cytokines through down-regulation of target proteins such as IL-1 receptor activated kinase 1 (IRAK1) and TNF receptor-associated factor 6 (TRAF6), which are the mediators for NF-κB activation (92,93). TLR4 signaling has a very important role in the up-regulation of inflammation and the release of inflammatory cytokines in animal models and also in in vitro cellular models of ALI (92,93). miR-146a mimic treatment suppressed LPS-mediated pro-inflammatory cytokines induction in normal alveolar rat macrophage cells and also suppressed inducible nitric oxide synthase (iNOS), favoring M2 macrophage phenotype and amelioration of ALI in mice lungs exposed to acid and LPS-treated macrophages (92). Therefore, miR-146a could be a therapeutic target in ALI. Recently, a study has shown that plasma levels of miRNA-146a and miRNA155 significantly increased in patients with severe sepsis and sepsis-induced ALI as compared to control subjects, and may be biomarkers predictive of mortality and treatment outcome of sepsis-induced ALI (94).
Conclusions
Research on the role of miRNAs in the pathogenesis, diagnosis and pathophysiology-based treatment options in ALI/ARDS is at its infancy. A substantial body of evidence suggests that indeed certain miRNAs are related to the pathophysiology of ALI/ARDS and may be interesting diagnostic biomarkers and therapeutic targets. However, more research is necessary before these discoveries are translated into clinical practice. In addition, translational experimental designs are needed to discover biomarker of ARDS as a clinicopathological entity, to identify patients sharing the same phenotype (i.e., ARDS with DAD).
Acknowledgements
Funding: The study was supported by Instituto de Salud Carlos III, FEDER EU Funding (FIS PI 15/01942).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Rubenfeld GD, Caldwell E, Peabody E, et al. Incidence and outcomes of acute lung injury. N Engl J Med 2005;353:1685-93. [Crossref] [PubMed]
- Ferguson ND, Fan E, Camporota L, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med 2012;38:1573-82. [Crossref] [PubMed]
- Travis WD, Costabel U, Hansell DM, et al. An official American Thoracic Society/European Respiratory Society statement: Update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med 2013;188:733-48. [Crossref] [PubMed]
- Katzenstein AL, Bloor CM, Leibow AA. Diffuse alveolar damage--the role of oxygen, shock, and related factors. A review. Am J Pathol 1976;85:209-28. [PubMed]
- Tomashefski JF Jr. Pulmonary pathology of acute respiratory distress syndrome. Clin Chest Med 2000;21:435-66. [Crossref] [PubMed]
- Thille AW, Vuylsteke A, Bersten A. Does the Berlin definition for acute respiratory distress syndrome predict the presence of diffuse alveolar damage? Intensive Care Med 2015;41:342-4. [Crossref] [PubMed]
- Cardinal-Fernández P, Bajwa EK, Dominguez-Calvo A, et al. The Presence of Diffuse Alveolar Damage on Open Lung Biopsy Is Associated With Mortality in Patients With Acute Respiratory Distress Syndrome: A Systematic Review and Meta-Analysis. Chest 2016;149:1155-64. [Crossref] [PubMed]
- Guerin C. DAD in nonresolving ARDS provides support for prolonged glucocorticoid treatment: a rebuttal. Intensive Care Med 2015;41:1166-7. [Crossref] [PubMed]
- Kao KC, Hu HC, Chang CH, et al. Diffuse alveolar damage associated mortality in selected acute respiratory distress syndrome patients with open lung biopsy. Crit Care 2015;19:228. [Crossref] [PubMed]
- Lorente JA, Cardinal-Fernandez P, Munoz D, et al. Acute respiratory distress syndrome in patients with and without diffuse alveolar damage: an autopsy study. Intensive Care Med 2015;41:1921-30. [Crossref] [PubMed]
- Villar J, Blanco J, Kacmarek RM. Current incidence and outcome of the acute respiratory distress syndrome. Curr Opin Crit Care 2016;22:1-6. [Crossref] [PubMed]
- Cross LJ, Matthay MA. Biomarkers in acute lung injury: insights into the pathogenesis of acute lung injury. Crit Care Clin 2011;27:355-77. [Crossref] [PubMed]
- Villar J, Slutsky AS. GOLDEN anniversary of the acute respiratory distress syndrome: still much work to do! Curr Opin Crit Care 2017;23:4-9. [Crossref] [PubMed]
- Terpstra ML, Aman J, van Nieuw Amerongen GP, et al. Plasma biomarkers for acute respiratory distress syndrome: a systematic review and meta-analysis*. Crit Care Med 2014;42:691-700. [Crossref] [PubMed]
- Blondonnet R, Constantin JM, Sapin V, et al. A Pathophysiologic Approach to Biomarkers in Acute Respiratory Distress Syndrome. Dis Markers 2016;2016:3501373.
- Bos LD, Weda H, Wang Y, et al. Exhaled breath metabolomics as a noninvasive diagnostic tool for acute respiratory distress syndrome. Eur Respir J 2014;44:188-97. [Crossref] [PubMed]
- Villar J, Perez-Mendez L, Espinosa E, et al. Serum lipopolysaccharide binding protein levels predict severity of lung injury and mortality in patients with severe sepsis. PLoS One 2009;4:e6818. [Crossref] [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597-610. [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3-14. [Crossref] [PubMed]
- Berezikov E, Chung WJ, Willis J, et al. Mammalian mirtron genes. Mol Cell 2007;28:328-36. [Crossref] [PubMed]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005;6:376-85. [Crossref] [PubMed]
- Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA sequences in human disease. Mutat Res 2012;731:14-9. [Crossref] [PubMed]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and function. Thromb Haemost 2012;107:605-10. [Crossref] [PubMed]
- Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol 2006;3:97-100. [Crossref] [PubMed]
- Liu J, Valencia-Sanchez MA, Hannon GJ, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 2005;7:719-23. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Mullany LE, Herrick JS, Wolff RK, et al. MicroRNA Seed Region Length Impact on Target Messenger RNA Expression and Survival in Colorectal Cancer. PLoS One 2016;11:e0154177. [Crossref] [PubMed]
- Reddemann K, Gola D, Schillert A, et al. Dysregulation of microRNAs in angioimmunoblastic T-cell lymphoma. Anticancer Res 2015;35:2055-61. [PubMed]
- van Schooneveld E, Wildiers H, Vergote I, et al. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res 2015;17:21. [Crossref] [PubMed]
- Zhang Q, Kandic I, Kutryk MJ. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem Biophys Res Commun 2011;405:42-6. [Crossref] [PubMed]
- Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol 2010;2:161-79. [PubMed]
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672-5. [Crossref] [PubMed]
- Cardinal-Fernández P, Ferruelo A, Esteban A, et al. Characteristics of microRNAs and their potential relevance for the diagnosis and therapy of the acute respiratory distress syndrome: from bench to bedside. Transl Res 2016;169:102-11. [Crossref] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009;136:642-55. [Crossref] [PubMed]
- O'Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A 2007;104:1604-9. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255-60. [Crossref] [PubMed]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta 2008;1779:471-8.
- Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta 2008;1779:697-701.
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 2006;66:7390-4. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Zhao H, Shen J, Medico L, et al. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One 2010;5:e13735. [Crossref] [PubMed]
- Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118-26. [Crossref] [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. [Crossref] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [Crossref] [PubMed]
- Etheridge A, Lee I, Hood L, et al. Extracellular microRNA: a new source of biomarkers. Mutat Res 2011;717:85-90. [Crossref] [PubMed]
- Weiland M, Gao XH, Zhou L, et al. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol 2012;9:850-9. [Crossref] [PubMed]
- Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009;19:439-48. [Crossref] [PubMed]
- Zhang X, Liu S, Hu T, et al. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009;50:490-9. [Crossref] [PubMed]
- Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128-36. [Crossref] [PubMed]
- Crawford M, Brawner E, Batte K, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun 2008;373:607-12. [Crossref] [PubMed]
- Rehman I, Evans CA, Glen A, et al. iTRAQ identification of candidate serum biomarkers associated with metastatic progression of human prostate cancer. PLoS One 2012;7:e30885. [Crossref] [PubMed]
- Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol 2015;35:69-77. [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009;9:581-93. [Crossref] [PubMed]
- Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 2006;20:1487-95. [Crossref] [PubMed]
- Muralidharan-Chari V, Clancy JW, Sedgwick A, et al. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010;123:1603-11. [Crossref] [PubMed]
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 2012;110:483-95. [Crossref] [PubMed]
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 2012;110:483-95. [Crossref] [PubMed]
- Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet 2010;3:484-8. [Crossref] [PubMed]
- Chen X, Liang H, Zhang J, et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012;22:125-32. [Crossref] [PubMed]
- Etheridge A, Gomes CP, Pereira RW, et al. The complexity, function and applications of RNA in circulation. Front Genet 2013;4:115. [Crossref] [PubMed]
- Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013;33:186-92. [Crossref] [PubMed]
- Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol 2013;228:1713-9. [Crossref] [PubMed]
- Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol 2016;36:301-12. [Crossref] [PubMed]
- Kim KM, Abdelmohsen K, Mustapic M, et al. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017.8. [PubMed]
- Quesenberry PJ, Aliotta J, Camussi G, et al. Potential functional applications of extracellular vesicles: a report by the NIH Common Fund Extracellular RNA Communication Consortium. J Extracell Vesicles 2015;4:27575. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet 2013;4:142. [Crossref] [PubMed]
- Angulo M, Lecuona E, Sznajder JI. Role of MicroRNAs in lung disease. Arch Bronconeumol 2012;48:325-30. [PubMed]
- Bhargava M, Wendt CH. Biomarkers in acute lung injury. Transl Res 2012;159:205-17. [Crossref] [PubMed]
- Benz F, Roy S, Trautwein C, et al. Circulating MicroRNAs as Biomarkers for Sepsis. Int J Mol Sci 2016;17:E78. [Crossref] [PubMed]
- Cai ZG, Zhang SM, Zhang Y, et al. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol 2012;90:37-43. [Crossref] [PubMed]
- Li W, Qiu X, Jiang H, et al. Downregulation of miR-181a protects mice from LPS-induced acute lung injury by targeting Bcl-2. Biomed Pharmacother 2016;84:1375-82. [Crossref] [PubMed]
- Fang Y, Gao F, Hao J, et al. microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2. Am J Transl Res 2017;9:1287-96. [PubMed]
- Xie T, Liang J, Liu N, et al. MicroRNA-127 inhibits lung inflammation by targeting IgG Fcgamma receptor I. J Immunol 2012;188:2437-44. [Crossref] [PubMed]
- Ying H, Kang Y, Zhang H, et al. miR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J Immunol 2015;194:1239-51. [Crossref] [PubMed]
- Li Q, Ge YL, Li M, et al. miR-127 contributes to ventilator-induced lung injury. Mol Med Rep 2017;16:4119-26. [Crossref] [PubMed]
- Vaporidi K, Vergadi E, Kaniaris E, et al. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 2012;303:L199-207. [Crossref] [PubMed]
- Cheng Y, Kuang W, Hao Y, et al. Downregulation of miR-27a* and miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation 2012;35:1308-13. [Crossref] [PubMed]
- Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007;179:5082-9. [Crossref] [PubMed]
- Mitchelson KR, Qin WY. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem 2015;6:162-208. [Crossref] [PubMed]
- Yu H, Lu Y, Li Z, et al. microRNA-133: expression, function and therapeutic potential in muscle diseases and cancer. Curr Drug Targets 2014;15:817-28. [Crossref] [PubMed]
- Wang Z, Ruan Z, Mao Y, et al. miR-27a is up regulated and promotes inflammatory response in sepsis. Cell Immunol 2014;290:190-5. [Crossref] [PubMed]
- Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Ann N Y Acad Sci 2010;1183:183-94. [Crossref] [PubMed]
- Schmidt WM, Spiel AO, Jilma B, et al. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun 2009;380:437-41. [Crossref] [PubMed]
- Vasilescu C, Rossi S, Shimizu M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One 2009;4:e7405. [Crossref] [PubMed]
- Roderburg C, Luedde M, Vargas Cardenas D, et al. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 2013;8:e54612. [Crossref] [PubMed]
- Wang W, Liu Z, Su J, et al. Macrophage micro-RNA-155 promotes lipopolysaccharide-induced acute lung injury in mice and rats. Am J Physiol Lung Cell Mol Physiol 2016;311:L494-506. [Crossref] [PubMed]
- Guo Z, Wen Z, Qin A, et al. Antisense oligonucleotide treatment enhances the recovery of acute lung injury through IL-10-secreting M2-like macrophage-induced expansion of CD4+ regulatory T cells. J Immunol 2013;190:4337-48. [Crossref] [PubMed]
- Vergadi E, Vaporidi K, Theodorakis EE, et al. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol 2014;192:394-406. [Crossref] [PubMed]
- Zeng Z, Gong H, Li Y, et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res 2013;39:275-82. [Crossref] [PubMed]
- Han Y, Li Y, Jiang Y. The Prognostic Value of Plasma MicroRNA-155 and MicroRNA-146a Level in Severe Sepsis and Sepsis-Induced Acute Lung Injury Patients. Clin Lab 2016;62:2355-60. [Crossref] [PubMed]


