The next generation of rapid point-of-care testing identification tools for ventilator-associated pneumonia
Introduction
Despite some advances in the understanding of ventilator-associated pneumonia (VAP) pathophysiology and the causing pathogens, VAP remains a major concern in the critical care setting because of its high incidence (1) and consequences in terms of mortality, morbidity and cost (2,3). VAP represents both a diagnostic and therapeutic challenge.
Clinical and radiologic diagnostic criteria have been shown to be inconsistent with autopsy findings (4) and simple laboratory tests such as C-reactive protein (CRP) and procalcitonin (PCT) are still not recommended, despite some promising results, because of concerns of limited sensitivity and specificity (5). The highest level of certainty for the diagnosis of VAP is thus achieved using “traditional” microbiological data, namely Gram stain examination which is a fast but inaccurate technique (6), and culture-based identification, obtained through a process that has only been slightly altered since it was first developed at the end of the nineteenth century. The main breakthrough in culture-based clinical microbiology was the recent widespread availability of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (7) that was shown to decrease the delay to get identification and susceptibility data by more than 10 h. Nevertheless, no significant impact was reported by this before-after study on patient outcomes, except intensive care unit (ICU) length of stay. Several polymerase chain reaction (PCR) kits have been developed or are still under development and could be used in the microbiological diagnosis of VAP, but are still not widely available, probably because of concerns regarding the lack of specificity, and increased cost compared to culture-based techniques (8).
Treating VAP is a difficult task, as initial antimicrobial therapy has to be appropriate and prompt (9,10), and changing the antimicrobial regimen after the first day does not seem to improve outcome (11). However, broad-spectrum antibiotic treatment must be used with caution, as the evidence for a direct association between antibiotics consumption and the emergence of multi-drug-resistant organisms (MDROs) cannot be neglected (12,13). Infections related to these bacteria are associated with high rate of inappropriate antibiotic treatment and worse outcomes (14).
Therefore, the need for nearly-instant diagnostic tools arises, and one of the potential methods to perform quick VAP diagnosis is the use of point-of-care testing (POCT) tools. The term POCT encloses any diagnostic tool performed “at the bedside” or close to it, without relying on central laboratory testing (15). POCT tools are now available in a large variety of settings and are vastly used in ICUs, the most commonly used are the measurement of blood glucose, hemoglobin concentration and arterial blood gas test. They tend to make essential diagnostic tests easily and quickly available, in particular by removing the time needed to package and transport the sample, identify the sample at the laboratory and transmit the result back to the ICU. Timely diagnosis can lead to quickly accurate treatment, which in turn can significantly improve patient outcome, particularly in the ICU setting. POCT tools sometimes also allow lower volumes of blood sample to obtain similar results. Concern remains regarding the reliability of these tools, and their results often need to be taken with caution, sometimes requiring traditional laboratory testing challenge.
Therefore, it seems logical to investigate the accuracy of POCT tools for the diagnosis of VAP. This narrative review aims to discuss the current data on POCT tools in the ICU setting, and its potential impact on near-future improvements and innovations.
Non-specific POCT tools: CRP and PCT
Although biomarkers are not recommended in the diagnosis of VAP (5), some of them are widely used in the clinical setting. Three biomarkers, including soluble triggering receptor expressed on myeloid cells type 1 (sTREM-1), CRP and PCT, were recently investigated for VAP diagnosis (16). However, these markers are not specific and increase in their concentration can be related to both infectious and non-infectious origins. To our knowledge, no POCT is available to this date for sTREM-1.
CRP has a reported sensitivity ranging from 56% to 88% and a specificity ranging from 86% to 91% (17,18), with large uncertainty regarding positivity cut-off (96 to 196 mg/L). CRP could nevertheless be used as a screening tool given its rather high specificity. Some authors also suggest using CRP as a prognostic marker in VAP, as a decrease in CRP during the course of treatment of VAP is associated with patient outcome. In a recent study by Póvoa et al. (19), survivors were patients who showed a decrease in CRP levels at day 4 of antimicrobial therapy of more than half the initial value, while patients who did not survive showed nearly no change in CRP level. There was also an association between adequate antibiotic therapy and a decrease in CRP level (P=0.029). POCT CRP has been available for a few years now, and several studies showed that POCT CRP can be useful for community-acquired pneumonia in outpatients (20), as it allows a reduction in antibiotics prescription without worsening outcome. A cost-effectiveness study also showed interesting results, encouraging the use of these tests in this setting (21). There is, however, no data regarding their interest for VAP.
PCT seems to have more interesting diagnostic value, with an estimated sensitivity between 78% to 100% and specificity between 75% and 97% (16). However, other authors reported lower diagnostic accuracy, with sensitivity as low as 41% (22) and specificity as low as 24% (23). PCT could be an interesting tool to discontinue antimicrobial therapy safely in the ICU according to recent studies (24,25), and as such could represent a potential candidate as a POCT tool. POCT to laboratory testing correlation seems to be sufficient (26), but there is no available data regarding its use for VAP, and few results are available for community-acquired pneumonia (27).
Automated microscopy as a POCT tool for VAP
Accelerate Diagnostics™ has been developing automated microscopy tools for many years, based on fluorescent in situ hybridization (FISH), allowing for both rapid pathogen identification and antibiotic susceptibility test (AST). The majority of available data come from positive blood culture, with promising results. Microbiological studies report overall sensitivity of 95.6% and specificity of 99.5% (respectively 95.6% and 99.1% for Gram-positive bacteria, 95.3% and 99.9% for Gram-negative bacteria and 100% and 98.9% for yeasts) (28), with a reported time to identification reduction of 23.5 h and time to AST reduction of 41.9 h compared to conventional microbiological techniques. Data for clinical settings are not yet available in large amount, but preliminary works like that of Kollef et al. (29) report a time to identification and AST of 10.2 (range, 8.3–11.5) h for the automated microscopy system vs. 51.4 (range, 48.0–54.6) h for conventional methods, with a projected impact on appropriate antibiotics change of 35.8 h and antimicrobial therapy de-escalation of 41.1 h.
Metzger et al. reported the use of this method to identify Staphylococcus aureus and Pseudomonas aeruginosa from broncho-alveolar lavage (BAL) samples taken from patients presenting with VAP (30). Species identification was successful in every sample and unaffected by the presence of a non-targeted specimen (Klebsiella pneumoniae). Positive concordance was reported as 13/16 for specimens containing target species above the diagnostic threshold, and 86/90 for specimens containing species under that threshold.
More recently, Douglas et al. reported the use of this technology as a microbiological surveillance method for VAP in ICU patients (31). This study used 73 samples collected from 33 patients, with a reported sensitivity of 100% (7/7) and specificity of 97% (64/66) compared to conventional microbiological culture. Interestingly, one of the false positive results for automated microscopy occurred in a patient that was diagnosed with VAP 2 days later on clinical criteria. The projected impact of this innovative technology on antibiotics prescription was a change in antibiotic regimen in 3/7 patients (43%) that would have occurred in about 5 h.
Since this technology uses fully automated sample preparation, FISH and AST, it could thus be easily used inside the ICU, with acceptable encumbrance similar to that of ABG POCT modules, and environment requirements (temperature, humidity) compatible with a clinical setting (32).
Multiplex PCR (MPCR): a promising diagnostic method and a POCT candidate
Over the last years, MPCR has been the subject of a steadily growing interest when it comes to microbiological identification in various clinical situations. The most recent kits not only allow pathogen identification, but can also reveal the presence of the most frequently encountered drug-resistance genes. Prior antimicrobial therapy does not influence PCR diagnostic accuracy. In addition, PCR-based methods allow for detection of non-bacterial pathogens. MPCR has been primarily developed for use with blood samples, and several data regarding this setting are available, including a recent randomized trial published by Banerjee et al. showing a significant decrease in broad-spectrum antimicrobial treatment for patients whose positive blood cultures were analyzed using PCR (44 vs. 56 h, P=0.01), as well as an increased narrow-spectrum antibiotic use (71 vs. 42 h, P=0.04) and a reduction in treatment of contaminants (11% vs. 25%, P=0.015) when compared to culture-based identification and AST, without significant difference in mortality, length of stay or cost (33). However, other randomized controlled trials, and meta-analyses reported less encouraging results (34). Several potential explanations could be suggested for this discrepancy, including the fact that blood cultures might not be the gold standard for blood stream infections, and the limited panel of tested bacteria and resistance genes tested in previous studies (35).
Jamal et al. studied the accuracy of the Unyvero Pneumonia Application (Curetis AG, Holzgerlingen, Germany) for the microbiological diagnosis of health care-associated pneumonia (36). In this analysis, performed in 49 patients, there was a reported agreement rate between MPCR and conventional culture of 63.3%, mostly due to MPCR detecting more organisms than culture. The Unyvero testing allowed for antimicrobial therapy regimen adjustment for 33 patients, with promising results on outcome, although the lack of a control group does not allow for an estimation of the impact on patient prognosis.
Baudel et al. investigated the accuracy of MPCR for the diagnosis of VAP and hospital-acquired pneumonia (HAP) in ICU patients (37). In this pilot study, the authors used the LightCycler 2.0 SeptiFast kit (Roche Diagnostics, Mannheim, Germany) that is only intended for use with blood samples, and PCR was not performed in real time. Among 65 patients with suspected pneumonia and 53 with confirmed pneumonia, MPCR allowed the identification of a pathogen in 66% of samples, vs. 23% for direct examination (P<0.001) and 40% for culture (P=0.01), and the identification rate was taken up to 82% after censoring the specimens containing species for which there was no probe in the MPCR kit. Interestingly, the pathogen identification rate was not influenced by ongoing antimicrobial therapy (66% vs. 64%, not significant). This kit does not allow resistance gene detection except for the detection of methicillin-resistant Staphylococcus aureus, and thus no data is available regarding this matter. Despite these promising results, this study raises serious concerns regarding colonization-related positive MPCR results, as 42% of the samples gathered from patients for whom the diagnosis of pneumonia was eventually ruled out came back positive.
To address this issue, Clavel et al. (38) interestingly chose to correlate the cycle threshold (Ct) obtained in real-time quantitative PCR with colony-forming units (CFU)/mL threshold obtained from conventional culture-based methods analyzed by nephelometry. Using this method, the authors report a sensitivity of 89.2% (range, 83.2–93.6%) and a specificity of 97.1% (range, 96.1–97.9%) using BAL samples, and 71.8% (range, 61.0–81.0%) sensitivity and 96.6% (range, 95.4–97.5%) using endotracheal aspirates (ETA). This work was a proof-of-concept study using various primers independently and thus no data are available regarding time to identification. The method could nevertheless be fully automated according to the authors.
Progress continues to be made on the miniaturization of PCR techniques. In a recent work, Pendleton et al. report the use of a palm-sized device weighing less than 100 g: MinION (Oxford Nanopore Technologies, Oxford, UK) (39). Contrary to standard MPCR methods that detect a predefined genetic sequence (e.g., 16S rRNA), this method allows for whole genome sequencing. In a few hours, the authors were able to obtain several thousand base-pair sequences that not only allowed for species identification, but also an accurate prediction of AST. MinION has also been successfully used with urine samples, allowing for identification for all studied samples and AST for 51 out of 55 samples in about 4 h (40). This method would not be bottlenecked by the “taxonomic bet” imposed by primers, although much additional work is needed to be able to automate the use of this tool in ICU setting. In its current state, the high cost of this device is also a serious setback for routine use.
To date, the major report regarding the use of MPCR as a POCT tool is a study by Kunze et al. using the Unyvero (Curetis AG, Holzgerlingen, Germany) kit (41). In this study, respiratory samples from 40 patients diagnosed with HAP were analyzed both by conventional culture-based tests and using the Unyvero MPCR. Results were obtained on average in 71 h (min–max: 37.2–217.8 h) using conventional methods vs. 6.5 h (min–max: 4.7–18.3 h) for MPCR. In “real-life” conditions, MPCR seem less reliable at the moment, as 10% of the sample analyses resulted in complete failure and another 30% resulted in partial failure. Among the successful or partially successful MPCR analyses, only half of the results were concordant with conventionally obtained results. Discordance was also of a concerning level when it came to predicting antibiotic resistances, probably because of the detection or resistance-related genes from resident species of the airways. This study calls for precaution regarding the use of MPCR inside the ICU, and would position this technique as an add-on to standard care, thus incurring additional cost that would have to be imbalanced with a strong benefit.
There are two major concerns regarding the routine use of MPCR as a POCT tool, contamination and colonization. Firstly, MPCR methods are known to be highly sensitive and require thorough precaution so as not to contaminate samples and produce false positive during manipulation. Laboratory personnel are well trained to this measure, but it would probably be completely otherwise in a clinical setting with partially trained staff working in the hurry of ICU situations. Secondly, thresholds to differentiate between infection and colonization of the airways are still the subject of controversies using conventional methods, and this concern would probably be emphasized by the use of MPCR, with uncertainty regarding the Ct considered as a positive result.
Other emerging methods for bedside VAP diagnosis
Exhalome analysis
One of the POCT tools that have been available for years is used on a daily basis for purposes completely distinct from healthcare, it is the detection of ethanol in exhaled breath. This principle could be used to diagnose VAP in ICU patients. Although research has been steadily active in this field, we still cannot use this easily available sample for microbiological diagnosis, but it could be the case in the near future. Mass spectrometry allows for real-time analysis of exhaled breath or volatile organic compounds (VOC) from various biological samples (including urine to detect urinary tract infections) (42). It is now possible to obtain a “breathprint” or exhalome in real-time (43), and it is easily conceivable to use fast detection of exhaled breath modifications as a VAP diagnostic tool just like we use modifications of urine composition detected by test strip as a diagnostic tool for urinary tract infections.
Since the first prototype reported in 1954, electronic noses (eNoses) have been the subject of many technological advances (44), allowing for miniaturization. Industry is now capable of offering handheld devices at the size of talkie-walkies like the Cyranose 320 (Sensigent, Baldwin Park, California, USA) that was used in a 2004 study by Hockstein et al. to assess the positive diagnosis of pneumonia with a reported prediction rate between 80% and 91.6% (45). These could make solid candidates as POCT tools for VAP.
In 2014, Bos et al. reported promising results for the use of eNose in VAP (46), with an area under the curve (AUC) of receiver operating characteristic (ROC) curve of 0.85 [95% confidence interval (CI), 0.69–1.00], not that different from that of the clinical pulmonary infection score (CPIS) (AUC, 0.89; 95% CI, 0.79–0.99) with a significant improvement when combining both diagnostic tools (AUC, 0.94; 95% CI, 0.86–1.00). The same group reported a feasibility study showing acceptable repeatability, no adverse event during the collection procedure and a low consumable cost (47).
In the study performed by Filipiak et al. (48), exhalome analysis allowed the authors to successfully detect the presence of pathogen-specific VOCs in samples non-invasively collected from mechanically ventilated patients with suspected VAP in a few hours. They also reported an observed concordance between the variations of the concentration of pathogen-specific VOCs in exhaled breath and the blood CRP level, suggesting a potential interest as a surveillance tool for measuring therapeutic response.
In another recent study, Schnabel et al. reported that a subset of 12 VOCs could be used to discriminate patients with VAP from control for whom that diagnosis was ruled out with a sensitivity of 75.8%±13.8% and a specificity of 73.0%±11.8% (ROC AUC, 0.87) in less than an hour (49).
Rapid chromogenic tests
Chromogenic tests have been used for the management of acute pharyngitis for more than a decade now (50), allowing for the selection of patients in need of treatment and thus promoting antibiotic stewardship in outpatients. These tests have a high specificity, and negative predictive value (51). In recent years, similar chromogenic tests have been developed in order to detect the production of extended-spectrum beta-lactamase (ESBL) by Enterobacteriaceae. These tests can detect ESBL and/or carbapenemase production in 30 to 120 min, with a sensitivity ranging from 80% to 95% and a specificity between 71% and 100% (52). Some tests designed to specifically detect carbapenemases are also under development, although at an early stage (53).
Recently, Garnier et al. reported the use of one of these tests, the β-LACTA™ test (BLT, Bio-Rad, Marnes-la-Coquette, France), in the early escalation or de-escalation of early antibiotic therapy in ICU patients diagnosed with infections caused by Enterobacteriaceae species (54). Among the 122 patients included in this analysis, 86 (70%) were diagnosed with pneumonia, and the majority of infections were hospital-acquired in this study (66, 54%). The use of BLT allowed for a more appropriate antibiotic regimen in the BLT-guided group (98% vs. 77%, P<0.01) with a significant reduction in time needed for antimicrobial therapy escalation [27 (range, 24–28) vs. 50.5 (range, 48–73) h, P<0.01] compared to the standard care control group.
Most of these tests were designed for use with bacterial colonies, but successful detection has been reported using fresh urine samples (55). Detection in respiratory samples appears to be possible according to another work by the same team (56). In this work, the authors report a sensitivity and specificity (and thus positive and negative predictive value) of 100%, using 126 bronchial aspirates containing ESBL producing Gram-negative bacterial strains above the 104 CFU/mL threshold. The main limitation regarding the use of this technique as a POCT test is the need for sample preparation—including digestion, haemolysis, centrifugation and incubation—that seem unfit for bedside clinical practice, despite only adding up to about an hour in this study. Automation of this process might be feasible using automated sample preparation instruments.
Conclusions
A long way ahead remains when it comes to moving microbiology from the laboratory to the bedside, but promising techniques are becoming widely available and could represent serious candidates for POCT tools (Figure 1). A comprehensive list of those techniques is proposed in Table 1, with data regarding time-to-identification, AST study mode and specifications.
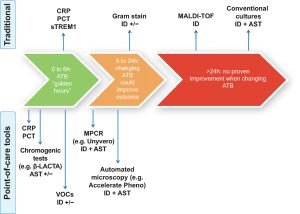
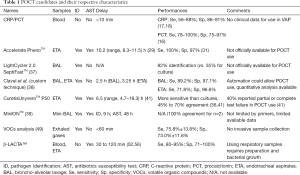
Full table
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Dudeck MA, Weiner LM, Allen-Bridson K, et al. National Healthcare Safety Network (NHSN) report, data summary for 2012, Device-associated module. Am J Infect Control 2013;41:1148-66. [Crossref] [PubMed]
- Kollef MH, Shorr A, Tabak YP, et al. Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest 2005;128:3854-62. [Crossref] [PubMed]
- Kollef MH, Hamilton CW, Ernst FR. Economic impact of ventilator-associated pneumonia in a large matched cohort. Infect Control Hosp Epidemiol 2012;33:250-6. [Crossref] [PubMed]
- Wunderink RG. Clinical criteria in the diagnosis of ventilator-associated pneumonia. Chest 2000;117:191S-194S. [Crossref] [PubMed]
- Kalil AC, Metersky ML, Klompas M, et al. Executive Summary: Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016;63:575-82. [Crossref] [PubMed]
- Blot F, Raynard B, Chachaty E, et al. Value of Gram Stain Examination of Lower Respiratory Tract Secretions for Early Diagnosis of Nosocomial Pneumonia. Am J Respir Crit Care Med 2000;162:1731-7. [Crossref] [PubMed]
- Mok JH, Eom JS, Jo EJ, et al. Clinical utility of rapid pathogen identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in ventilated patients with pneumonia: A pilot study. Respirology 2016;21:321-8. [Crossref] [PubMed]
- Torres A, Lee N, Cilloniz C, et al. Laboratory diagnosis of pneumonia in the molecular age. Eur Respir J 2016;48:1764-78. [Crossref] [PubMed]
- Iregui M, Ward S, Sherman G, et al. Clinical Importance of Delays in the Initiation of Appropriate Antibiotic Treatment for Ventilator-Associated Pneumonia. Chest 2002;122:262-8. [Crossref] [PubMed]
- Dupont H, Mentec H, Sollet JP, et al. Impact of appropriateness of initial antibiotic therapy on the outcome of ventilator-associated pneumonia. Intensive Care Med 2001;27:355-62. [Crossref] [PubMed]
- Luna CM, Vujacich P, Niederman MS, et al. Impact of BAL Data on the Therapy and Outcome of Ventilator-Associated Pneumonia. Chest 1997;111:676-85. [Crossref] [PubMed]
- Vibet MA, Roux J, Montassier E, et al. Systematic analysis of the relationship between antibiotic use and extended-spectrum beta-lactamase resistance in Enterobacteriaceae in a French hospital: a time series analysis. Eur J Clin Microbiol Infect Dis 2015;34:1957-63. [Crossref] [PubMed]
- Vernaz N, Huttner B, Muscionico D, et al. Modelling the impact of antibiotic use on antibiotic-resistant Escherichia coli using population-based data from a large hospital and its surrounding community. J Antimicrob Chemother 2011;66:928-35. [Crossref] [PubMed]
- Barbier F, Lisboa T, Nseir S. Understanding why resistant bacteria are associated with higher mortality in ICU patients. Intensive Care Med 2016;42:2066-9. [Crossref] [PubMed]
- Larsson A, Greig-Pylypczuk R, Huisman A. The state of point-of-care testing: a european perspective. Ups J Med Sci 2015;120:1-10. [Crossref] [PubMed]
- Palazzo SJ, Simpson T, Schnapp L. Biomarkers for ventilator-associated pneumonia: Review of the literature. Heart Lung 2011;40:293-8. [Crossref] [PubMed]
- Póvoa P, Coelho L, Almeida E, et al. C-reactive protein as a marker of infection in critically ill patients. Clin Microbiol Infect 2005;11:101-8. [Crossref] [PubMed]
- Ramirez P, Garcia MA, Ferrer M, et al. Sequential measurements of procalcitonin levels in diagnosing ventilator-associated pneumonia. Eur Respir J 2008;31:356-62. [Crossref] [PubMed]
- Póvoa P, Martin-Loeches I, Ramirez P, et al. Biomarkers kinetics in the assessment of ventilator-associated pneumonia response to antibiotics - results from the BioVAP study. J Crit Care 2017;41:91-7. [Crossref] [PubMed]
- Point-of-care CRP testing in the diagnosis of pneumonia in adults. Drug Ther Bull 2016;54:117-20. [Crossref] [PubMed]
- Cals JWL, Ament AJHA, Hood K, et al. C-reactive protein point of care testing and physician communication skills training for lower respiratory tract infections in general practice: economic evaluation of a cluster randomized trial. J Eval Clin Pract 2011;17:1059-69. [Crossref] [PubMed]
- Duflo F, Debon R, Monneret G, et al. Alveolar and Serum ProcalcitoninDiagnostic and Prognostic Value in Ventilator-associated Pneumonia. Anesthesiology 2002;96:74-9. [Crossref] [PubMed]
- Luyt CE, Combes A, Reynaud C, et al. Usefulness of procalcitonin for the diagnosis of ventilator-associated pneumonia. Intensive Care Med 2008;34:1434-40. [Crossref] [PubMed]
- Tang H, Huang T, Jing J, et al. Effect of Procalcitonin-Guided Treatment in Patients with Infections: a Systematic Review and Meta-Analysis. Infection 2009;37:497-507. [Crossref] [PubMed]
- Bouadma L, Luyt CE, Tubach F, et al. Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet 2010;375:463-74. [Crossref] [PubMed]
- Dupuy AM, Chevrier Q, Olejnik Y, et al. Analytical evaluation of point-of-care procalcitonin (PCT) and clinical performances in an unselected population as compared with central lab PCT assay. Clin Chem Lab Med 2017;55:e167-71. [Crossref] [PubMed]
- Masiá M, Padilla S, Ortiz de la Tabla V, et al. Procalcitonin for selecting the antibiotic regimen in outpatients with low-risk community-acquired pneumonia using a rapid point-of-care testing: A single-arm clinical trial. PLoS One 2017;12:e0175634. [Crossref] [PubMed]
- Tesic V, Hill B, Love N, et al. Accelerate PhenoTM System for the Identification and Susceptibility (ID/AST) of Pathogens in Positive Blood Cultures and Impact on Time to Results and Workflow. ASM Microbe 2017. Available online: http://acceleratediagnostics.com/wp-content/uploads/2017/06/ASM-Microbe-2017-U.-Chicago.pdf
- Kollef M, Burnham C, Fuller B. Automated microscopy for rapid directed antibiotic treatment of sepsis. ISICEM 2017. Available online: http://www.intensive.org/admin/upload/abstract/1079190576/P/P398.pdf
- Metzger S, Frobel RA, Dunne WM. Rapid simultaneous identification and quantitation of Staphylococcus aureus and Pseudomonas aeruginosa directly from bronchoalveolar lavage specimens using automated microscopy. Diagn Microbiol Infect Dis 2014;79:160-5. [Crossref] [PubMed]
- Douglas IS, Price CS, Overdier KH, et al. Rapid Automated Microscopy for Microbiological Surveillance of Ventilator-associated Pneumonia. Am J Respir Crit Care Med 2015;191:566-73. [Crossref] [PubMed]
- Specifications. Accelerate Diagnostics. Available online: http://acceleratediagnostics.com/products/accelerate-pheno-system/specifications/
- Banerjee R, Teng CB, Cunningham SA, et al. Randomized Trial of Rapid Multiplex Polymerase Chain Reaction–Based Blood Culture Identification and Susceptibility Testing. Clin Infect Dis 2015;61:1071-80. [Crossref] [PubMed]
- Warhurst G, Maddi S, Dunn G, et al. Diagnostic accuracy of SeptiFast multi-pathogen real-time PCR in the setting of suspected healthcare-associated bloodstream infection. Intensive Care Med 2015;41:86-93. [Crossref] [PubMed]
- Nseir S, Povoa P. Multipathogen real-time PCR system adds benefit for my patients: no. Intensive Care Med 2015;41:531-3. [Crossref] [PubMed]
- Jamal W, Al Roomi E. Evaluation of Curetis Unyvero, a multiplex PCR-based testing system, for rapid detection of bacteria and antibiotic resistance and impact of the assay on management of severe nosocomial pneumonia. J Clin Microbiol 2014;52:2487-92. [Crossref] [PubMed]
- Baudel JL, Tankovic J, Dahoumane R, et al. Multiplex PCR performed of bronchoalveolar lavage fluid increases pathogen identification rate in critically ill patients with pneumonia: a pilot study. Ann Intensive Care 2014;4:35. [Crossref] [PubMed]
- Clavel M, Barraud O, Moucadel V, et al. Molecular quantification of bacteria from respiratory samples in patients with suspected ventilator-associated pneumonia. Clin Microbiol Infect 2016;22:812.e1-812.e7. [Crossref] [PubMed]
- Pendleton KM, Erb-Downward JR, Bao Y, et al. Rapid Pathogen Identification in Bacterial Pneumonia Using Real-time Metagenomics. Am J Respir Crit Care Med 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Schmidt K, Mwaigwisya S, Crossman LC, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 2017;72:104-14. [Crossref] [PubMed]
- Kunze N, Moerer O, Steinmetz N, et al. Point-of-care multiplex PCR promises short turnaround times for microbial testing in hospital-acquired pneumonia--an observational pilot study in critical ill patients. Ann Clin Microbiol Antimicrob 2015;14:33. [Crossref] [PubMed]
- Smith D, Španěl P. Selected ion flow tube mass spectrometry (SIFT-MS) for on-line trace gas analysis. Mass Spectrom Rev 2005;24:661-700. [Crossref] [PubMed]
- Martinez-Lozano Sinues P, Zenobi R, Kohler M. Analysis of the Exhalome. Chest 2013;144:746-9. [Crossref] [PubMed]
- Wilson AD, Baietto M. Applications and Advances in Electronic-Nose Technologies. Sensors 2009;9:5099-148. [Crossref] [PubMed]
- Hockstein NG, Thaler ER, Torigian D, et al. Diagnosis of Pneumonia With an Electronic Nose: Correlation of Vapor Signature With Chest Computed Tomography Scan Findings. Laryngoscope 2004;114:1701-5. [Crossref] [PubMed]
- Bos LD, Martin-Loeches I, Kastelijn JB, et al. The volatile metabolic fingerprint of ventilator-associated pneumonia. Intensive Care Med 2014;40:761-2. [Crossref] [PubMed]
- Bos LD, Wang Y, Weda H, et al. A simple breath sampling method in intubated and mechanically ventilated critically ill patients. Respir Physiol Neurobiol 2014;191:67-74. [Crossref] [PubMed]
- Filipiak W, Beer R, Sponring A, et al. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: a prospective pilot study. J Breath Res 2015;9:016004. [Crossref] [PubMed]
- Schnabel R, Fijten R, Smolinska A, et al. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci Rep 2015;5:17179. [Crossref] [PubMed]
- Humair JP, Revaz SA, Bovier P, et al. Management of Acute Pharyngitis in Adults: Reliability of Rapid Streptococcal Tests and Clinical Findings. Arch Intern Med 2006;166:640-4. [Crossref] [PubMed]
- Stefaniuk E, Bosacka K, Wanke-Rytt M, et al. The use of rapid test QuikRead go® Strep A in bacterial pharyngotonsillitis diagnosing and therapeutic decisions. Eur J Clin Microbiol Infect Dis 2017;36:1733-8. [Crossref] [PubMed]
- Poirel L, Fernández J, Nordmann P. Comparison of Three Biochemical Tests for Rapid Detection of Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae. J Clin Microbiol 2016;54:423-7. [Crossref] [PubMed]
- Mancini S, Kieffer N, Poirel L, et al. Evaluation of the RAPIDEC® CARBA NP and β-CARBA® tests for rapid detection of Carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 2017;88:293-7. [Crossref] [PubMed]
- Garnier M, Rozencwajg S, Pham T, et al. Evaluation of early antimicrobial therapy adaptation guided by the BetaLACTA® test: a case-control study. Crit Care 2017;21:161. [Crossref] [PubMed]
- Gallah S, Decré D, Genel N, et al. The β-Lacta Test for Direct Detection of Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae in Urine. J Clin Microbiol 2014;52:3792-4. [Crossref] [PubMed]
- Gallah S, Benzarara Y, Tankovic J, et al. β LACTA test performance for detection of extended-spectrum β-lactamase-producing Gram-negative bacilli directly on bronchial aspirates samples: a validation study. Clin Microbiol Infect 2017. [Epub ahead of print]. [Crossref] [PubMed]


