Switch-on the LAMP to spot Zika
Zika fever is caused by an arbovirus (a virus transmitted by insects) belonging to the Flavivirus genus of the Flaviviridae family, like dengue tick-borne encephalitis, west Nile virus and yellow fever, just to name a few. The insect vector of the disease is the female mosquito of Aedes aegypti, which originated in Africa. Aedes albopictus (A. albopictus) (the tiger mosquito from Asia) could also transmit the Zika virus, as it already does for dengue and Chikungunya virus, which is another arbovirus sharing similar vectors and geographical distribution with dengue and Zika virus. The Zika virus was detected for the first time in a monkey in Uganda in 1947. The first human cases appeared in Africa and then in Asia, but for decades the infection spread unnoticed. The current epidemic broke out in 2007 in Micronesia (Yap Islands in the Pacific Ocean). In 2013 a large epidemic occurred in French Polynesia, which then spread to other islands in the Pacific. The Zika virus was detected for the first time in the northwest of Brazil in May 2015 and quickly spread to neighbouring countries. As of January 2017, almost all Latin American and Caribbean countries have reported active Zika virus circulation. Zika virus of the Asian lineage coming from Brazil emerged in the continental USA (Florida and Texas) and spread to Cape Verde now challenging Africa again. Europe experienced several imported cases of Zika infection from travellers. The vector A. albopictus is present in Southern Europe, but no autochthonous cases of Zika virus transmission have been reported so far. Several excellent reviews have been published on the current Zika virus epidemic and the reader is encouraged to take advantage of them for a more comprehensive understanding of the disease, for example (1,2). A mosquito becomes infected with Zika virus during a blood meal when it bites a person with circulating Zika virus (urban circulation). The virus multiplies in the mosquito without affecting vector viability and is transmitted to another victim in the next blood meal. The viremic phase lasts few weeks after the bite and, during this time, the person can infect other mosquitoes or spread the infection vertically to newborns and horizontally through sexual intercourse or transfusion. Most people infected with the virus develop mild symptoms, if any, which do not require hospitalization. These symptoms are similar to those of Dengue and Chikungunya infections, which share the same vectors and geographical distribution, thus making the diagnosis difficult without a specific test (3,4). Complications of Zika infection include the Guillain-Barré syndrome, a progressive ascending paralysis, which could affect the respiratory muscles. Pregnant women can transmit the virus to their unborn children and this leads to microcephaly with severe neurological complications (5,6).
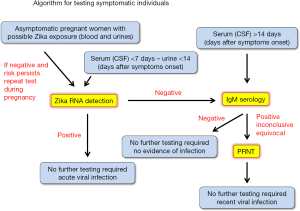
Standard laboratory diagnosis of Zika virus infection includes virus isolation, molecular detection of the viral genome and serology. Following the recommendations of the Centre for Disease Control of the Unites States and the World Health Organization an algorithm for the diagnosis of Zika virus has been designed to guide laboratory analysis. As simplified in Figure 1, suspected infected individuals within a week from the onset of symptoms are assayed for the presence of viral RNA in blood. Prolonged shedding in the urines allows a longer window of opportunity to detect the virus (about 2 weeks). Given the overlap of symptoms and sharing of vector and location, simultaneous detection of Dengue and Chikungunya RNA should be considered. A positive result is definitive for acute viral infection. A negative result should be followed by IgM serology and eventually confirmed by the plaque reduction neutralization test (PRNT). Positivity is indicative of a recent infection. In the case of pregnant women at risk (travel to endemic areas, sexual exposure) repeated testing during pregnancy is highly recommended.
Nunya Chotiwan, Connie D. Brewster and Joel Rovnak from the College of Veterinary Medicine and Biomedical Sciences, Colorado State University in Fort Collins recently published their results on the detection of Zika virus by loop-mediated isothermal amplification (LAMP) of viral RNA (7). A large network of international collaborators where involved in the study, which included countries endemic for Zika virus such as Brazil and Nicaragua, which provided valuable samples. The LAMP assay they propose allows the lineage-specific detection of Zika virus RNA in cultured infected cells, mosquitoes, virus-spiked samples of human secretions (blood, plasma, saliva, urine, and semen) and in infected patients’ biofluids. The LAMP for Zika RNA is conducted in a one-step reaction taking advantage of the reverse transcriptase and strand-displacement activity of Bst DNA polymerase. Sensitivity is in the range of a standard RT-PCR and specificity is Zika lineage-specific and not reactive for closely related Dengue and Chikungunya viruses. No extraction of viral RNA is required in most cases, although poorly preserved samples and/or specimens with low viral RNA may still require a step of RNA extraction/concentration to maximize the chances of success. Furthermore, the limit of detection of LAMP varies a lot between different biofluids, with semen spiked with viral RNA being the worse. This latter aspect is particularly important and should be investigated in more detail. To note, several reports have shown that Flavivirus such as west Nile virus and tick-borne encephalitis appear to be associated to the erythrocyte fraction of whole blood (8,9). Indeed, prolonged Zika virus viremia exceeding two months was detected in whole blood (10,11). These findings have obvious implications for the risk of virus transmission by blood transfusion, but also for the methodology of specimen preparations for diagnosis that usually process serum or plasma rather than whole blood. Further investigation in this aspect is ongoing.
LAMP is a very sensitive nucleic acid amplification assay (12). Usually two or three set of primers, with the addition of two loop primers to accelerate the reaction, target a specific DNA region for highly specific amplification. The DNA target sequence is amplified at a constant temperature of 60–65 °C by a polymerase with high strand displacement activity. Addition of a reverse transcriptase activity allows amplification of RNA. The amount of DNA produced in LAMP is considerably higher than in a PCR-based amplification reaction and can be visually detected by the turbidity caused by a by-product of amplification (magnesium pyrophosphate precipitate). The reaction can be followed in real-time also by fluorescence using intercalating dyes and amplifications of the same template are directly comparable for quantification purposes, although limited to end-point comparisons because the amplification is not linear as it is in PCR. LAMP-based assays for infectious disease diagnosis are getting more and more popular because they provide specificity and sensitivity in the range of standard RT PCR without the need of a thermal cycler. Furthermore, LAMP has been observed to be more resistant than PCR to inhibitors present in biofluids such as blood allowing minimal processing of the samples. The combination of low cost of operation and detection combined with the high specificity of the reaction make LAMP a promising tool to develop rugged point-of-care tests (POCT) for resource-limited settings. Major drawbacks of the technique are related to the number of primers required for the amplification of a limited fragment of DNA: although they provide high specificity, they also need careful optimization to avoid primer-primer interactions, therefore limiting the choice of the target region, as well as making multiplex reactions unfeasible.
The development of the LAMP assay for Zika virus is part of an international effort to provide POCT for a number of viral diseases affecting the developing world (13). Each aspect of the algorithm shown in Figure 1 has been considered for POCT or accelerated/automated processing. In this respect, a rapid acceleration in the field has been certainly prompted by the current Zika virus epidemic. A number of POCT for Zika virus RNA amplification in LAMP format have been recently proposed (14-19). LAMP appears more adaptable to POCT than PCR, although the amplification technology is far more advanced and widely implemented (see for example the list of Zika virus emergency use authorizations from CDC at https://www.fda.gov/MedicalDevices/Safety/EmergencySituations/ucm161496#zika). To complete the picture, NS1 antigen-capture is being proposed for viremia detection in POCT formats as an alternative to nucleic-acid detection (20,21). Serology is dominated by low cost mono-parametric strip tests, which are low cost and easy to use. However, there is a growing demand for syndrome-based multi-parametric devices with an electronic interface to process and archive the data (22,23). Finally, the most laborious and time-consuming PRNT is also being automated for rapid analysis (24,25). It is clear that from this wide number of approaches only few will eventually survive. In this respect it is highly recommended to adhere as close as possible to the World Health Organization “ASSURED” criteria, which describe the ideal characteristics of a diagnostic test that can be used at all levels of the health care system: affordable, sensitive, specific, user-friendly, rapid, equipment-free, and delivered to those who need them (26).
Conclusion and outlook
The work of Chotiwan and collaborators, together with that of several other groups, is particularly important because they not only provide the coordinates of the LAMP reaction for Zika virus detection, but also analysed carefully the reaction conditions in different biofluids. Optimization of the whole process, from extraction to detection, is critical for a successful diagnostic assay because each step adds to the complexity of the reaction and contributes to determining the final cost of the test. This becomes indispensable particularly when designing POCT assays to be deployed in resource-limited settings.
Acknowledgements
Work on flaviviruses is supported by the Beneficientia Stiftung, Vaduz Lichtenstein and by the FLAVIPOC and SEVARE projects from the Regione FVG of Italy.
Footnote
Conflicts of Interest: The authors have collaboration with Euroclone SpA and ProXentia Srl for the development of POCT for Flavivirus detection.
References
- Baud D, Gubler DJ, Schaub B, et al. An update on Zika virus infection. Lancet 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Weaver SC, Costa F, Garcia-Blanco MA, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res 2016;130:69-80. [Crossref] [PubMed]
- Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev 2016;29:487-524. [Crossref] [PubMed]
- Waggoner JJ, Pinsky BA. Zika Virus: Diagnostics for an Emerging Pandemic Threat. J Clin Microbiol 2016;54:860-7. [Crossref] [PubMed]
- Calvet G, Aguiar RS, Melo AS, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis 2016;16:653-60. [Crossref] [PubMed]
- Mlakar J, Korva M, Tul N, et al. Zika Virus Associated with Microcephaly. N Engl J Med 2016;374:951-8. [Crossref] [PubMed]
- Chotiwan N, Brewster CD, Magalhaes T, et al. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci Transl Med 2017;9:eaag0538. [Crossref] [PubMed]
- Rios M, Daniel S, Chancey C, et al. West Nile virus adheres to human red blood cells in whole blood. Clin Infect Dis 2007;45:181-6. [Crossref] [PubMed]
- Caracciolo I, Bassetti M, Paladini G, et al. Persistent viremia and urine shedding of tick-borne encephalitis virus in an infected immunosuppressed patient from a new epidemic cluster in North-Eastern Italy. J Clin Virol 2015;69:48-51. [Crossref] [PubMed]
- Lustig Y, Mendelson E, Paran N, et al. Detection of Zika virus RNA in whole blood of imported Zika virus disease cases up to 2 months after symptom onset, Israel, December 2015 to April 2016. Euro Surveill 2016.21. [PubMed]
- Murray KO, Gorchakov R, Carlson AR, et al. Prolonged Detection of Zika Virus in Vaginal Secretions and Whole Blood. Emerg Infect Dis 2017;23:99-101. [Crossref] [PubMed]
- Notomi T, Mori Y, Tomita N, et al. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 2015;53:1-5. [Crossref] [PubMed]
- Baba M, Vidergar N, Marcello A. Virological point-of-care testing for the developing world. Future Virology 2014;9:595-603. [Crossref]
- Lee D, Shin Y, Chung S, et al. Simple and Highly Sensitive Molecular Diagnosis of Zika Virus by Lateral Flow Assays. Anal Chem 2016;88:12272-8. [Crossref] [PubMed]
- Song J, Mauk MG, Hackett BA, et al. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal Chem 2016;88:7289-94. [Crossref] [PubMed]
- Tian B, Qiu Z, Ma J, et al. Attomolar Zika virus oligonucleotide detection based on loop-mediated isothermal amplification and AC susceptometry. Biosens Bioelectron 2016;86:420-5. [Crossref] [PubMed]
- Wang X, Yin F, Bi Y, et al. Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J Virol Methods 2016;238:86-93. [Crossref] [PubMed]
- Ganguli A, Ornob A, Yu H, et al. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed Microdevices 2017;19:73. [Crossref] [PubMed]
- Calvert AE, Biggerstaff BJ, Tanner NA, et al. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP). PLoS One 2017;12:e0185340. [Crossref] [PubMed]
- Bosch I, de Puig H, Hiley M, et al. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci Transl Med 2017.9. [PubMed]
- Afsahi S, Lerner MB, Goldstein JM, et al. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens Bioelectron 2018;100:85-8. [Crossref] [PubMed]
- Marcello A, Sblattero D, Cioarec C, et al. A deep-blue OLED-based biochip for protein microarray fluorescence detection. Biosens Bioelectron 2013;46:44-7. [Crossref] [PubMed]
- Tagliabue G, Faoro V, Rizzo S, et al. A label-free immunoassay for Flavivirus detection by the Reflective Phantom Interface technology. Biochem Biophys Res Commun 2017;492:558-64. [Crossref] [PubMed]
- Maistriau M, Carletti T, Zakaria MK, et al. A method for the detection of virus infectivity in single cells and real time: Towards an automated fluorescence neutralization test. Virus Res 2017;237:1-6. [Crossref] [PubMed]
- Wilson HL, Tran T, Druce J, et al. Neutralization Assay for Zika and Dengue Viruses by Use of Real-Time-PCR-Based Endpoint Assessment. J Clin Microbiol 2017;55:3104-12. [Crossref] [PubMed]
- Mabey D, Peeling RW, Ustianowski A, et al. Diagnostics for the developing world. Nat Rev Microbiol 2004;2:231-40. [Crossref] [PubMed]


