Prognostic and predictive biomarkers post curative intent therapy
Introduction
Lung cancer is the second most commonly diagnosed cancer in both men and women with an estimated 222,500 new cases expected to be diagnosed in 2017, and an estimated 155,870 deaths, more than any other cancer site (1). A minority of lung cancers (only 16%) are diagnosed at a localized stage which is the only window of disease that is potentially curable with surgery (2). Nonetheless, 5-year survival ranges between 30–50% for lung cancers diagnosed at stage I or II, thus despite early detection and curative resection, a significant proportion of patients will die from recurrent disease within 5 years (1,2).
Furthermore, most lung cancers are diagnosed when the disease has metastasized, increasing the difficulty to treat which parallels the 4% 5-year survival rates in the most advanced stages (1). In fact, studies have shown that good clinical outcomes are limited to lung cancer patients who are diagnosed before blood vessel invasion and before the disease metastasizes to the regional lymph nodes or has locally advanced (3,4). For this reason, much effort has focused on improving the rates of early diagnosis through improved imaging technology and adaption of screening programs.
The survival heterogeneity among early-stage lung cancers is a clinical challenge even with multi-modality approaches. Systemic adjuvant chemotherapy (ACT) provides relatively small gains in benefit, with only 5% improvement in 5-year survival (5). Equally confusing are the data demonstrating post-operative radiation therapy (PORT) in patients with early stage lung cancer. Furthermore, utilization of multi-modality treatments is guided only by basic clinical criteria like tumor size or nodal involvement (6).
This review will focus on the role of predictive and prognostic markers in the post-curative intent setting with an emphasis on three key areas: (I) prognostic signatures to stratify patients at high- and low-risk for recurrence; (II) predictive markers for conventional cytotoxic chemotherapy and (III) the role of targeted therapies in early stages of disease.
Historical perspective on lung cancer standards of practice
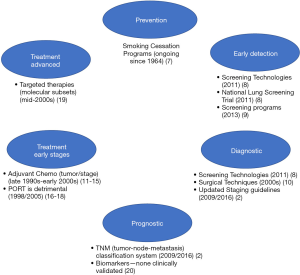
Various advances spanning the past several decades have evolved the standards of practice for lung cancer (see Figure 1).
Lung cancer is largely preventable given most cases are due to tobacco smoking (1), thus large efforts related to smoking cessation programs spanning several decades have been made to reduce the incidence of new lung cancer cases (7). The National Lung Screening Trial (NLST), a prospective randomized controlled trial (RCT) with over 53,000 patients demonstrated lung cancer screening with low-dose computed tomography (LDCT) was more effective in reducing lung cancer deaths in high-risk groups (8) than traditional radiography, thus leading to implementation of lung cancer screening guidelines in 2013 (9) and providing evidence for early diagnosis as a key to improved survival. Advancements in technology have continued to facilitate similar progress in the diagnosis and accurate staging of lung cancers, with minimally invasive procedures such as endobronchial ultrasounds, mediastinoscopy and video-assisted thoracoscopic surgery (10). Stage of lung cancer at diagnosis is a critical element in determining prognosis and appropriate treatment strategy, therefore many of these imaging and surgical advancements have facilitated more accurate determination of disease spread, and consequently have also facilitated continual improvements in staging guidelines (2).
A subset of early-stage lung cancers are at high-risk for recurrence, thus the utility of chemotherapy after complete resection for early stages has been investigated in several large RCTs, systematic reviews and meta-analyses, with the hope of demonstrating an improvement in survival with ACT. Disappointingly, improvements in 5-year overall survival (OS) are modest ranging between 4–15% (5,11-14) with the use of various platinum-based regimens (without post-operative radiotherapy). Based on the accumulating evidence from the past few decades, guidelines issued by American Society of Clinical Oncology and Cancer Care Ontario (15) and National Comprehensive Cancer Network (NCCN) (6) recommend adjuvant cisplatin-based ACT as standard of care for stage II and III NSCLC, however, the use of ACT in stage I has been more controversial. Although no universal standard can be applied, evidence would suggest that survival advantages of ACT may be reserved for stage I patients with larger tumors (≥4 cm; stage IB) (16). Other considerations when weighing the benefits of ACT may include performance status, quality of life, toxic effects of chemotherapy and comorbidities, as many of patients included in the clinical trial setting are enriched for mild or no comorbidities and good performance status.
Meta-analyses and systemic reviews continue to provide mixed evidence for the effect of PORT on survival for early stage tumors (N0–1 disease), and stage III (N2) disease (6,17-19). First established in 1998, recently published systemic reviews confirm PORT leads to increased risk of death (18%) and overall reduction in survival, regardless of stage or nodal status (18). Studies with modern radiotherapy techniques using linear accelerators, such as stereotactic radiation therapy or stereotactic body radiation therapy (SBRT) are currently under investigation and may help to clarify the role of PORT in post-curative NSCLC. Definitive local therapy, either alone or in combination with chemotherapy may be considered for inoperable early stages, as well as resectable stage IIIA, as well as stage IV cancer with limited metastatic sites (6).
Precision medicine in oncology has witnessed an explosion in recent years, with identification of molecularly-defined subtypes of disease and development of targeted therapies (e.g., EGFR, ALK, ROS1, T790M etc.). The identification of targetable driver mutations and their association with better prognosis (when treated with targeted therapies) have been observed only in the advanced disease setting (20) (stage IV) and the data have not yet been replicated in the adjuvant setting. However, studies investigating these markers in early stage lung cancer patients are underway.
Prognostic and predictive markers in the post-curative intent setting
Prognostic and predictive biomarkers
In the post-curative intent setting of lung cancer, there are two areas of potential application of biomarkers. First, identifying patients that are at increased risk for recurrence after curative resection and therefore clinical management of these patients would include more aggressive strategies, including adjuvant treatment. Such markers are considered prognostic, able to distinguish clinical outcomes regardless of therapy selection (21), and most well-known examples of such clinical prediction models are Oncotype DX® or MammaPrint® utilized in breast cancer. Secondly, once adjuvant therapy is determined to be necessary for a patient, biomarkers that guide selection of specific therapies, or predictive biomarkers, are also of high interest for early stages of lung cancer. Predictive biomarkers guide patient selection for specific treatment options and have demonstrated their utility in enriching patients that are likely to derive substantial clinical benefit verses those that may not (21). In lung cancer, many prognostic and predictive markers and signatures have been assessed, however, to date, only predictive markers (e.g., EGFR, ALK) have been clinically validated and incorporated into practice guidelines for advanced stages (6). Selected biomarkers utilized in clinical practice, have been studied or are under active investigation for early stages of lung cancer are summarized in Table 1.

Full table
Due to their potentially important role in cancer management, the development of guidelines for tumor marker studies, both prognostic and predictive, have been outlined previously (21,27). However, most marker studies to date have not met the suggested requirements around study design, statistical analyses, or assay development and methods. Therefore, there is a relative shortage of validated prognostic and predictive markers that are well-supported. The lack of validated tumor markers for early-stage lung cancer does not imply the lack of need, rather is likely a result of the focus of developing and improving lung cancer standards of practice in the past several decades.
Given the high rates of recurrence and mortality for NSCLC there is a clear need for delineating molecular characteristics of tumors that have more aggressive tumor biology reflecting a potential for a tumor to metastasize. In an analysis of a previous clinical decision tool utilized to select breast cancer patients at high-risk for recurrence, Tian and colleagues concluded that the multigene signature encompasses genes whose biological properties are related to the six hallmarks of cancer (evading apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, limitless replicative potential, tissue invasion and metastasis and sustained angiogenesis) as defined by Hanahan and Weinberg (28,29). In the pursuit of identifying such molecular characteristics for lung cancer, it is important to confirm these molecular changes occur early during tumorigenesis, as well as determine whether these characteristics apply across clinically relevant subtypes, such as smoking status and molecular or histological subtypes.
Prognostic
In lung cancer, there is much debate regarding the optimal application of ACT. Several large studies and meta-analyses have determined that ACT confers increases in 5-year survival rates, between 4–15%, for resected stage II–III lung cancers (2). However, selection of patients for ACT in high-risk stage I (IB) and low risk stage II (IIA), is not standardized. It is not clear whether Stage IB patients characterized by larger tumors (≥4 cm) that have not metastasized to the regional lymph nodes, or stage IIA patients which are smaller in size but have positive lymph nodes, should have ACT. Currently, only clinical factors such as stage and other risk factors for prognosis, such as extent of resection and positive margins, along with patient discussion and clinical intuition are used to decide for or against ACT in these settings, where guidelines are less clear. Consequently, it is of great interest to develop prognostic signatures that provide insight into the tumor biology to help stratify patients with lowest risk where ACT and related toxicities can be avoided, and identify patients that are at high-risk for recurrence, where ACT should be pursued.
Prognostic multigene signatures
Many prognostic multigene signatures derived mostly through genome-wide mRNA expression arrays to stratify NSCLC patients either categorically (e.g., low- or high-risk) or as a continuous score for recurrence have been developed (30-38) and several previous reviews also summarize these data (39-41). A recent large-scale meta-analysis compared and evaluated the prognostic performance of 42 published gene signatures in a large collection of diverse (heterogeneous survival outcomes, histology and tumor stage) datasets (42) which included 1,927 NSCLC patients. The comprehensive analysis utilized various statistical models to summarize their performance and found that approximately 59% or 25 of the studied signatures (number of genes ranged between 4–452) were prognostic for survival after adjustment for clinical factors. The meta-estimated hazard ratios (95% CI) for predicted high-risk groups ranged between 1.25–1.72 for adenocarcinoma-based signatures, and 1.20–1.41 for squamous cell carcinoma-based signatures, and notably, there was some but little overlap amongst the genes. As these authors conclude, the signatures with strongest statistical significance for predicting patient outcome deserve further assessment for feasibility of measurement of genes with assays performed in a clinical laboratory improvement amendments (CLIA) laboratory environment and that utilize FFPE samples, as well as selection of signatures with more feasible number of genes, all of which are challenges faced with mRNA-based approaches.
Despite an abundance of validated multigene signatures, including several that performed robustly in an independent meta-analysis, few have emerged as clinically validated prognosticators. Most of these studies involve a multi-stage process of signature discovery through analysis of gene expression data (historically microarray platforms) and hierarchical clustering or risk modeling in independent institutional patient cohorts where clinical outcomes are known. Once a signature or gene expression profile is identified in the training cohort, the assay is tested retrospectively in multiple rounds of independent validation cohorts, some of which are publicly available data sets [e.g., The Cancer Genome Atlas (TCGA)]. Lastly, to complete the clinical validation the signature is tested in a prospective RCT against current clinicopathological criteria, in the case of lung cancer, tumor size, stage and nodal status. To date, however, the majority of multigene signatures have not made it past validation in independent data sets.
There has been much debate regarding the reproducibility of mRNA expression platforms (43,44) and besides technological limitations (e.g., fresh-frozen tissue vs. formalin-fixed paraffin-embedded samples), biases (22) and statistical approaches (45), several other reasons can be speculated for the lack of fully validated prognostic signatures in lung cancer. As lung cancer subtyping moves from histological to genomic classifications, the failure to validate signatures may be due to the need for different signatures according to clinically relevant groupings, for example, EGFR-mutated (46), KRAS-mutated (23), or now by immunophenotypes (24). Population of patients being studied may be very heterogeneous, for example, mixed treatment populations or too wide of the clinical stages (early vs. late) or molecular subtypes. Lastly, translation of validated signatures into clinically useful tools requires the development of a signature that is adaptable to more common clinical samples (FFPE), CLIA environment-friendly assays and incorporates s a feasible number of genes (<100) (42).
The search for prognostic signatures has also expanded to utilize large-scale gene expression analyses with updated technologies that are more reflective of current knowledge of the genome, and may include interrogation of the non-coding portions of the genome, as well as interrogation of the methylome. This contrasts many of the historical profiling approaches which focused solely on the coding genome, thus combining protein-coding and non-coding genes may be a more comprehensive assessment to identify molecular signatures. In addition, these platforms may be more adaptable to utilization of clinical samples (e.g., FFPE) and to CLIA-certified environments. Using the RNA-Seq platform, Shukla and colleagues identified a four-gene signature which included protein-coding genes (RHOV, CD109 and FRRS1), as well as a long noncoding RNA gene (lncRNA), LINC00941, and stratified OS in clinical and mutation subsets (47). This signature statistically significantly stratified stage I lung adenocarcinomas for OS and metastasis-free survival (HR =2.78, 95% CI: 1.91–11.13, P<0.001; HR =3.30, 95% CI: 2.89–13.45, P<0.001, respectively) utilizing large TCGA and institutional cohorts. Notably, the four-gene signature also successfully stratified OS in EGFR-mutant and wildtype patients, thus providing potential utility in identification of EGFR-mutant patients that would benefit from intensification of targeted therapy strategies (47).
Other attempts to utilize the non-coding genome for prognostic purposes include several studies examining microRNA (miRNA) expression profiles to stratify patients that are at high-risk for relapse (48-50). miRNA are small, non-coding RNA molecules that can regulate gene expression that effect signaling, growth, differentiation and transformation, and several studies indicate their potential role as biomarkers for disease and progression (48). A study controlling for many of the common technical challenges when measuring miRNA levels (e.g., platform, specimen type), found significant differences in miRNA expression in therapy-naive tumor resection samples from patients that relapsed within 2 years to those that did not relapse within 3 years. By utilizing each patient as their own control to compare levels of miRNA expression in tumor cells and surrounding normal tissues, the authors found dysregulation of a specific set of miRNA in the environment around the tumor in patients’ tumors that recurred compared to those that did not (48). Additionally, multi-omic approaches for the development of prognostic classifiers have also been investigated. For instance, a signature comprised of protein-coding (XPO1, BRCA1, HIF1a and DLC1), non-coding (miR-21) and HOXA9 promoter methylation improved the statistical significance of identifying high-risk therapy naïve stage I lung adenocarcinomas than only utilizing coding or non-coding expression data alone (HR =10.2; P=3×10−5) (25,49,50). Although expansion of the technological approaches allows for a more comprehensive assessment of the tumor landscape is promising, these signatures are also still in preliminary stages of testing and have yet to be clinically validated.
To date, only a few multigene prognostic signatures have been commercialized and now undergoing clinical testing in prospective RCTs for their utility in prognostication of early stage lung cancers (26). The first, known as myPlan® Lung Cancer (Myriad, Salt Lake City, UT) is a multigene signature incorporating a cell cycle progression (CCP) score assessed through expression status (RNA extracted from FFPE and assayed by qRT-PCR) of 31 genes involved in the cell cycle, normalized to 15 housekeeping genes and incorporates pathological stage into the final risk categorization algorithm to provide a numerical score (51). Development of the assay was carried out using publicly available microarray datasets (Director’s Consortium cohort) and validation using independent institutional datasets (52).
Several studies in solid tumors, including lung (37,51), prostate (53) and breast cancer (54), have demonstrated the status of genes within the CCP cycle as an underlying measure of the aggressiveness of patient’s tumors regardless of molecular classifications and therefore an important prognostic tool, given the circuitry is the gateway to uncontrolled cell proliferation, an important hallmark of cancer (29). The 46-gene panel comprising the CCP score has been shown to be an independent predictor of 5-year risk of mortality in four multinational published studies (51,55-57). These data clearly demonstrated that the final prognostic algorithm which incorporates the CCP score and pathological stage provided more significant stratification power for lung cancer mortality risk than using the CCP score alone. The validated algorithm demonstrated that early stage lung cancer patients with a high myPlan Lung Cancer molecular prognostic score (mPS) were nearly twice as likely to succumb to their disease within 5 years compared to patients with a low mPS (average risk estimates were 35% vs. 18%) (52,55,56).
As a final validation of the myPlan Lung Cancer mPS, two parallel prospective registries are accruing patients to determine the effect of the test on treatment decisions of surgeons and oncologists (NCT02121925 and NCT02121899), as well as asses the disease-free survival (DFS). A completion date at the end of 2018 is planned (58).
The second commercialized assay, Pervenio™ Lung RS platform (Life Technologies, West Sacramento, CA) incorporates a risk score model developed based on expression levels measured by quantitative PCR utilizing FFPE non-squamous NSCLC tissue samples. The 14-gene signature (includes three reference genes) was identified by Cox proportional risk modeling in a large training set of early stage non-squamous NSCLC. The target genes include signaling components of oncogenic pathways known to be altered in lung cancer pathogenesis (e.g., ERBB3), DNA repair mechanisms (BRCA1) and cell cycle regulation (CDC6, CDK2AP1), among other genes involved in other various cellular processes. The validation set utilized was a masked cohort of 433 patients with stage I non-squamous NSCLC and over a 1,000 patients with stage I–III non-squamous NSCLC from the China Clinical Trials Consortium (CCTC) (34). The assay stratifies patients into three categorizations and Kaplan-Meier analysis demonstrated the 5-year OS was 71.4% (95% CI: 60.5–80) in low-risk, 58.3% (48.9–66.6) in intermediate-risk, and 49.2% (42.2–55.8) in high-risk patients (P trend =0.0003) for the first cohort and similar findings for the CCTC cohort (34). When compared with other clinical factors (histology, tumor size, histology, etc.) a multivariate analysis showed that the signature was significantly predictive of mortality (high-risk HR =2.04; P=0.0016) and subsequent validation studies found minimal bias of the prognostic signature (59).
A large, multicenter, randomized prospective trial to evaluate the effect of ACT in patients with completely resected stage I non-squamous NSCLC identified as high- risk by the Pervenio™ Lung RS assay (NCT01817192) is underway. Primary and secondary outcomes are OS and DFS, and study completion is expected in 2021 (58). Preliminary clinical performance of the assay was recently published in a small, single institution prospective study of 100 stage I–IIA non-squamous NSCLC tested with the Pervenio assay. Woodard and colleagues estimated 5-year DFS was 48.9% among high-risk patients who did not elect chemotherapy, 93.8% among untreated molecular low-risk patients, compared to 91.7% in high-risk patients who did elect chemotherapy (P=0.004) (60). Despite the small sample size and lack of randomization, this data provides promising preliminary evidence supporting a role of a prognostic tool for molecular stratification of early stage NSCLC that should undergo ACT.
Of note, a separate 15-gene signature developed from 133 patients in the JBR.10 trial (RCT of adjuvant vinorelbine/cisplatin versus observation alone) whose (frozen) tumor samples were assayed by microarray profiling has also been validated. The assay categorizes early stage NSCLC into low- and high-risk of recurrence after surgery, and when applied to patients that received ACT, treatment significantly prolonged survival of the high-risk patients, however was not beneficial to the low-risk group (HR =0.33; 95% CI: 0.17–0.63; P=0.0008 and HR =3.67; 95% CI: 1.22–11.06; P=0.021, respectively) (32).
Immunological milieu for prognosis
Immune escape, the underlying mechanisms malignant tumors evolve to evade host immune responses, plays an essential role in progression of tumors. Therapeutic targeting of the immune checkpoint pathways [e.g., programmed death-1 (PD-1)] is standard treatment for newly diagnosed PD-L1+ advanced NSCLC. The relationship between up-regulation of inhibitory receptors (PD-1) and ligands (PD-L1) of immune checkpoint pathways in early stages of NSCLC with survival is debatable, whereas the data suggesting the prognostic role of immune factors reflecting active host immune responses [e.g., tumor infiltrating lymphocytes (TILs)] has been more consistent. For instance, Tsao and colleagues (61) assessed the prognostic role of PD-L1 from tumor sections from three of the pivotal ACT trials (IALT, JBR.10 and CALB 9633) and found that PD-L1 expression correlated with squamous histology, intense lymphocytic infiltrate and KRAS mutations, however was not prognostic for survival. In contrast, studies have shown a higher degree of intratumoral TILs or FoxP3 positive regulatory TILs, as well as intense tumor lymphocytic infiltration associated with reduced likelihood of disease recurrence and improved survival, respectively, in resected samples from patients with early-stage NSCLC (24,62,63). The clinical relevance of measuring host immune factors during early stages of NSCLC may be important for supplementing TNM staging, as Bremnes and colleagues (64) developed an immuno-score classifying CD8+ T cells in the tumor stroma of NSCLC patients into three categories and showed this score had a prognostic impact within pathological stages (65).
Circulating biomarkers for disease recurrence
The detection of circulating tumor material, including circulating tumor- or cell-free-DNA (ctDNA or cfDNA) has potential for widespread applications, including non-invasive tumor biopsies for treatment decisions, as well preliminary data suggesting ctDNA following surgery or therapy with curative intent is prognostic for disease recurrence of various solid tumors (66-68). A recent study assessing KRAS and EGFR alleles, in pre- and post-surgery plasma samples of NSCLC patients by competitive allele-specific TaqMan PCR (CAST-PCR) (66). Compared to normal controls at 30 days after surgery, five patients who recurred within 4 months had significantly higher circulating cfDNA (P<0.001), whereas six patients who recurred after 4 months (P=0.207) and five patients without recurrence (P=0.901) demonstrated significantly lower circulating cfDNA. Therefore, analysis of circulating tumor DNA after local therapy with curative intent might provide a feasible strategy for identifying patients likely to relapse, and therefore identify patients for adjuvant treatment. As technology improves for non-invasive mutation detection for treatment decisions in the advanced setting, the application of these non-invasive techniques for detection of disease after curative resection will likely be explored further.
Predictive
Once therapy is deemed necessary for NSCLC, an additional area of interest would be the identification of predictive markers for customizing therapy selections. Despite the therapeutic effect of conventional cytotoxic chemotherapy regimens, responses vary and treatments are not without their toxic side effects, thus the role of pharmacogenomics, single nucleotide polymorphisms (SNPs) or expression status of markers relevant to the mechanism of action for cytotoxic chemotherapies are being investigated as strategies to refine treatment selection. Furthermore, molecular subtyping of lung cancer and the role of targeted therapies have had significant impact on extending survival in the advanced stages of patients with lung cancer, however, the role of these therapies in early settings of disease remain widely unknown.
Biomarkers of response to conventional chemotherapy
Cisplatin-based chemotherapy is the backbone of adjuvant treatment in lung cancer, often partnered with other cytotoxics such as vinorelbine or taxanes. Several studies examining the predictive role of biomarkers have been investigated, usually in retrospective analyses, based on the mechanism of action of these cytotoxic therapies. For example, platinum agents induce bulky adducts in the DNA of tumor cells, initiating DNA repair responses, predominantly nucleotide excision repair (NER) pathway as an attempt to maintain DNA integrity (69). Thus, components of the DNA repair pathways, such as excision repair cross-complementation group 1 or ERCC1, have been studied extensively for their potential role in resisting the effects of DNA damaging agents. Although not considered “targeted” therapies, cytotoxic therapies do have cellular “targets” (often multiple cellular targets), thus many studies to identify predictive markers for cytotoxic agents, such as β-tubulin class III (TUBB3) for vinorelbine and ribonucleotide reductase M1 (RRM1) for gemcitabine, have been studied for many solid tumors (70), however their predictive role, if any, were found only in retrospective single arm studies. Importantly, many cytotoxic therapies have multiple mechanisms of action, and therefore tying a single gene or protein biomarker to the effects of these therapies may not be an optimal approach.
One of the most widely-pursued predictive biomarkers for platinum therapies in the adjuvant setting was ERCC1, a rate-limiting component of the NER response pathway, however multiple large retrospective studies have not been able to validate its predictive role for the adjuvant setting. Initially, Olaussen et al. demonstrated ERCC1 negativity in stage I–III NSCLC patients was predictive of chemotherapy efficacy (P=0.009 for interaction), Bepler and colleagues also demonstrated a positive treatment effect on OS in ERCC1 negative patients (HR =0.73, 95% CI: 0.55–0.96; P=0.02) (71,72). However, a retrospective re-analysis of tumors collected from two large adjuvant trials (LACE and IALT) was not able to validate the predictive effect of ERCC1 status, even after testing sixteen antibodies (73). It is hypothesized that the lack of reproducibility is likely due to varying batches of antibodies (none were able to distinguish four ERCC1 protein isoforms) and cutoffs, differences in technology platforms (RT-PCR, IHC) as well as interinstitutional variations in specimen collection. In addition to RNA and protein expression, the presence of SNPs has also been widely explored due to data suggesting certain codon polymorphisms impair translation and/or stability of the protein, thereby affecting response to platinum therapy (69). These data have also not been translated into fully utilized clinical tools.
Role of targeted and immune therapies in earlier stages
Targeted therapies in the adjuvant setting have been proven to transform adjuvant care for gastrointestinal stromal tumors (GIST) and breast cancer, with imatinib and tamoxifen or trastuzumab, respectively (74). However, the presence and role of driver mutations in earlier stages of lung cancer is not clear. Preliminary studies examining the effect of targeted therapies in the adjuvant setting were initiated prior to the identification of molecular drivers and were done in unselected populations. Therefore, a better understanding of lung pathogenesis would precipitate the utilization of targeted therapies in early settings.
Previous trials of EGFR targeted therapies in the adjuvant setting showed conflicting data regarding whether earlier treatment with TKIs is beneficial, most likely a result of selection criteria that did not focus on EGFR-sensitizing mutations. The NCIC CTG BR.19 trial randomized patients to gefitinib or placebo for 2 years with OS as a primary endpoint (75). There was no difference in DFS or OS at 6.3 years. A subset analysis of 15 patients with EGFR-mutated NSCLC (retrospectively assayed), demonstrated no DFS or OS benefit (seven received gefitinib, eight on placebo). Authors noted that the EGFR mutation rate was very low at 4% and hypothesized this mutation may occur as a later event in NSCLC pathogenesis. Another early trial, RADIANT (76) prospectively enrolled EGFR-positive (IHC/FISH and mutation) NSCLC patients randomized to erlotinib or placebo for 2 years with a primary endpoint of DFS. Adjuvant erlotinib did not prolong DFS in the overall population, however in the EGFR-mutated subset (16.5% of the cohort), erlotinib extended median DFS (46.4 vs. 28.5 months; HR =0.61; P=0.0391), however hierarchical clustering rendered this endpoint as non-significant. Additionally, the single-arm SELECT trial (77), a prospective trial which enrolled EGFR mutation positive stage I-IIIA NSCLC patients treated with 2 years of erlotinib after completion of standard chemotherapy and/or radiation. With a median follow-up of 3 years, the 2-year DFS was 90% (97% stage I, 73% stage II and 92% stage III). Interestingly, 24 patients recurred (2 during erlotinib treatment and 22 after stopping erlotinib) with a median time to recurrence of 12 months after stopping erlotinib. Fifteen of the recurrent patients underwent repeat biopsy and only one patient acquired EGFR T790M resistance mutation. Of the recurrent patients, many were re-treated with erlotinib and continued treatment (2–42 months), thus an important observation from this study is that generally these patients appear to remain sensitive to EGFR TKIs.
Lastly, the results of the highly-anticipated CTONG 1104 trial, the first RCT comparing gefitinib to cisplatin/vinorelbine (1:1) in completely resected stage II–IIIA NSCLC patients with N1/N2 disease and EGFR (L858R/del19) mutation positive were recently presented (78). Encouragingly, the data showed adjuvant gefitinib significantly prolonged DFS compared with the chemotherapy arm (28.7 months, 95% CI: 24.9–32.5 vs. 18.0 months, 95% CI: 13.6–22.3; HR =0.60; 95% CI: 0.42–0.87; P=0.005). Importantly, less toxicities and improved quality of life were observed on the gefitinib arm. Based on these results and possibly results from future large-scale prospective trials, EGFR-TKIs will continue to gain acceptance as potential treatment options in the adjuvant setting for early-stage EGFR-mutation positive NSCLC.
Studies to determine the role of targeted therapies applied in earlier settings of disease are also underway for other molecular subtypes, include ALK-rearranged NSCLC. To determine whether plausible, the true prognostic impact of ALK status was investigated in a recent retrospective analysis (79) comparing outcomes of various molecular subgroups of patients. A trend demonstrating ALK+ NSCLC had inferior relapse-free survival compared to EGFR+ NSCLC patients (HR =1.6, 95% CI: 0.9–2.8; P=0.104), after excluding patients that received adjuvant TKI.
Current large-scale efforts to determine the true role of genomic profiling and targeted therapies in early stages are underway, including the “Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trial”, or ALCHEMIST (NCT02194738, NCT02193282, NCT02201992) and ADUARA trial for osimertinib in EGFR-mutation positive patients (NCT02511106) (58). The cooperative ALCHEMIST study consists of three protocols: screening and the EGFR- and ALK-treatment trials. The screening trial will enroll up to 8,000 patients with stage 1–3 non-squamous NSCLC, where blood and tumor samples will be collected and sequenced with advanced genomic profiling, before or after surgical resection, and before or after ACT. Patients with EGFR or ALK mutations will be referred to the treatment arms which are double-blind, placebo-controlled trials with patients randomized (1:1) to placebo vs. erlotinib or crizotinib after standard chemotherapy. Duration of treatment with targeted therapies will be up to 2 years. Planned enrollment for these trials are 410 and 378 patients, respectively, with OS as a primary endpoint (80).
The role of the immune checkpoint pathway, PD-1 and associated immunotherapies are also being investigated in the adjuvant setting. Due to the lack of consensus for predictive biomarkers in the treatment setting of advanced disease, the design of trials for the adjuvant setting remain biomarker agnostic in terms of eligibility, however treatment interaction with biomarker status will be evaluated as part of key trials. Currently, PD-L1 positivity (which often tracks well with tumor mutation burden) and MSI status are utilized as biomarkers for pembrolizumab in advanced disease, however application of immunotherapies based on these molecular subtypes earlier in the disease course remains to be determined and are in the very early stages of clinical testing. The “Pembrolizumab (MK-3475) Versus Placebo for Patients with Early Stage NSCLC After Resection and Completion of Standard Adjuvant Therapy”, or the PEARLs (81), is an international adaptive design triple-blinded, placebo-controlled and randomized (1:1) study to evaluate pembrolizumab after surgery and standard chemotherapy with DFS as a primary endpoint, and will consider treatment effect across PD-L1 (programmed death ligand 1) subgroups. Randomization of 1,380 patients is planned. Additional studies assessing immunotherapy after resection include the ANVIL study, an open-label nivolumab versus observation trial (NCT02595944), BR31 trial, assessing durvulamab in a phase III double blind placebo controlled randomized study in resected NSCLC (NCT02273375) and lastly, a study evaluating the efficacy of an immunotherapy combination consisting of durvalumab and tremelimumab (CTLA-4), which recently demonstrated high levels of clinical activity in advanced disease settings, after standard adjuvant therapy (NCT03130764). Consideration of immunotherapy in earlier disease states will depend heavily on optimal patient selection criteria, since only a small proportion of patients obtain durable clinical benefit from these agents. Additional refinement of schedules, dosages and duration of therapy would also be important given the current cost of these therapies and potential for less-frequent dosing schedules (82).
Conclusions
The standards of practice in patients with early stage lung cancer are evolving. Improvements in cure rates have focused on early diagnosis, molecular subtypes and targeted therapies in advanced settings and improving surgical techniques. Through these advancements, the field is now poised to improve upon low cure rates through CT screening as well as the identification of patient selection tools to stratify patients by low and high-risk of recurrence. Risk modeling is attractive due to its success in other solid tumors (e.g., breast cancer) and large RCT are underway to validate in lung cancer. Historically we have tried to use single biomarkers to predict response and customize therapy, but that field is still largely exploratory with recent lack of success (e.g., ERCC1). It also seems unlikely that one single biomarker can predict for response for complicated mechanisms of action (platinum, DNA repair pathway). Now that numerous targeted therapies have been approved in the metastatic setting, this will facilitate the utilization in the early stage patient population. The role of targeted therapies is also of high interest, given that they have been important for other adjuvant settings, breast cancer or GIST, for example. Ongoing and planned studies will help define whether we will be able to incorporate risk models, biomarkers and targeted therapy in the curative patient population.
Acknowledgements
None.
Footnote
Conflicts of Interest: ES Kim: Consulting from Celgene; Boehringer Ingelheim; Eli Lilly; AstraZeneca. R Feldman has no conflicts of interest to declare.
References
- American Cancer Society. Cancer Facts & Figures 2017. Accessed 17th April 2017. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Wisnivesky JP, Henschke C, McGinn T, et al. Prognosis of Stage II non-small cell lung cancer according to tumor and nodal status at diagnosis. Lung Cancer 2005;49:181-6. [Crossref] [PubMed]
- Wang J, Chen J, Chen X, et al. Blood vessel invasion as a strong independent prognostic indicator in non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2011;6:e28844. [Crossref] [PubMed]
- Burdett S, Pignon JP, Tierney J, et al. Adjuvant chemotherapy for resected early-stage non-small cell lung cancer. Cochrane Database Syst Rev 2015.CD011430. [PubMed]
- NCCN Guidelines. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology. Version 5.2017. Accessed 16th March 2017.
- Koop CE, Luoto J. "The Health Consequences of Smoking: Cancer," overview of a report of the Surgeon General. Public Health Rep 1982;97:318-24. [PubMed]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Wender R, Fontham ET, Barrera E Jr, et al. American Cancer Society lung cancer screening guidelines. CA Cancer J Clin 2013;63:107-17. [Crossref] [PubMed]
- American Cancer Society. Early Detection, Diagnosis and Staging. In: Tests for Non-Small Cell Lung Cancer. 2016. Available online: https://www.cancer.org/cancer/non-small-cell-lung-cancer/detection-diagnosis-staging.html
- Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351-60. [Crossref] [PubMed]
- Chemotherapy in non-small cell lung cancer: a meta-analysis using updated data on individual patients from 52 randomised clinical trials. Non-small Cell Lung Cancer Collaborative Group. BMJ 1995;311:899-909. [Crossref] [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [Crossref] [PubMed]
- Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. [Crossref] [PubMed]
- Pisters KM, Evans WK, Azzoli CG, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I-IIIA resectable non small-cell lung cancer guideline. J Clin Oncol 2007;25:5506-18. [Crossref] [PubMed]
- McShane LM, Hayes DF. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol 2012;30:4223-32. [Crossref] [PubMed]
- Strauss GM, Herndon JE 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. [Crossref] [PubMed]
- Burdett S, Stewart L. PORT Meta-analysis Group. Postoperative radiotherapy in non-small-cell lung cancer: update of an individual patient data meta-analysis. Lung Cancer 2005;47:81-3. [Crossref] [PubMed]
- Burdett S, Rydzewska L, Tierney J, et al. Postoperative radiotherapy for non-small cell lung cancer. Cochrane Database Syst Rev 2016;9:Cd002142. [PubMed]
- Postoperative radiotherapy in non-small-cell lung cancer: systematic review and meta-analysis of individual patient data from nine randomised controlled trials. PORT Meta-analysis Trialists Group. Lancet 1998;352:257-63. [Crossref] [PubMed]
- McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97:1180-4. [Crossref] [PubMed]
- Tian S, Roepman P, Van't Veer LJ, et al. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark Insights 2010;5:129-38. [PubMed]
- Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768-77. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816-24. [PubMed]
- Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol 2007;25:5562-9. [Crossref] [PubMed]
- Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 2010;28:4417-24. [Crossref] [PubMed]
- Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 2006;355:570-80. [Crossref] [PubMed]
- Kratz JR, Van den Eeden SK, He J, et al. A prognostic assay to identify patients at high risk of mortality despite small, node-negative lung tumors. JAMA 2012;308:1629-31. [Crossref] [PubMed]
- Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 2012;379:823-32. [Crossref] [PubMed]
- Van Laar RK. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med Genomics 2012;5:30. [Crossref] [PubMed]
- Chen DT, Hsu YL, Fulp WJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011;103:1859-70. [Crossref] [PubMed]
- Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577-86. [Crossref] [PubMed]
- Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst 2010;102:464-74. [Crossref] [PubMed]
- Zhu CQ, Tsao MS. Prognostic markers in lung cancer: is it ready for prime time? Transl Lung Cancer Res 2014;3:149-58. [PubMed]
- Bunn PA Jr, Kim ES. Improving the Care of Patients With Stage IB Non-Small-Cell Lung Cancer: Role of Prognostic Signatures and Use of Cell Cycle Progression Biomarkers. Clin Lung Cancer 2015;16:245-51. [Crossref] [PubMed]
- Tang H, Wang S, Xiao G, et al. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann Oncol 2017;28:733-40. [Crossref] [PubMed]
- Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer 2007;7:545-53. [Crossref] [PubMed]
- Potti A, Mukherjee S, Petersen R, et al. Retraction: A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 2011;364:1176. [Crossref] [PubMed]
- Eklund AC, Szallasi Z. Correction of technical bias in clinical microarray data improves concordance with known biological information. Genome Biol 2008;9:R26. [Crossref] [PubMed]
- Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 2009;106:2824-8. [Crossref] [PubMed]
- Planck M, Isaksson S, Veerla S, et al. Identification of transcriptional subgroups in EGFR-mutated and EGFR/KRAS wild-type lung adenocarcinoma reveals gene signatures associated with patient outcome. Clin Cancer Res 2013;19:5116-26. [Crossref] [PubMed]
- Starmans MH, Pintilie M, Chan-Seng-Yue M, et al. Integrating RAS status into prognostic signatures for adenocarcinomas of the lung. Clin Cancer Res 2015;21:1477-86. [Crossref] [PubMed]
- Yan X, Jiao SC, Zhang GQ, et al. Tumor-associated immune factors are associated with recurrence and metastasis in non-small cell lung cancer. Cancer Gene Ther 2017;24:57-63. [Crossref] [PubMed]
- Shukla S, Evans JR, Malik R, et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. J Natl Cancer Inst 2016.109. [PubMed]
- Edmonds MD, Eischen CM. Differences in miRNA expression in early stage lung adenocarcinomas that did and did not relapse. PLoS One 2014;9:e101802. [Crossref] [PubMed]
- Akagi I, Okayama H, Schetter AJ, et al. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res 2013;73:3821-32. [Crossref] [PubMed]
- Robles AI, Arai E, Mathe EA, et al. An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma Based on mRNA, microRNA, and DNA Methylation Biomarkers. J Thorac Oncol 2015;10:1037-48. [Crossref] [PubMed]
- Robles AI, Harris CC. Integration of multiple "OMIC" biomarkers: A precision medicine strategy for lung cancer. Lung Cancer 2017;107:50-8. [Crossref] [PubMed]
- Zheng Y, Bueno R. Commercially available prognostic molecular models in early-stage lung cancer: a review of the Pervenio Lung RS and Myriad myPlan Lung Cancer tests. Expert Rev Mol Diagn 2015;15:589-96. [Crossref] [PubMed]
- Wistuba II, Behrens C, Lombardi F, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res 2013;19:6261-71. [Crossref] [PubMed]
- Myriad myPlan. What does myPlan Lung Cancer measure? Accessed 1st July 2017. Available online: https://myplanlungcancer.com/what-does-myplan-lung-cancer-measure/
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [Crossref] [PubMed]
- Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics 2008;1:11. [Crossref] [PubMed]
- Bueno R, Hughes E, Wagner S, et al. Validation of a molecular and pathological model for five-year mortality risk in patients with early stage lung adenocarcinoma. J Thorac Oncol 2015;10:67-73. [Crossref] [PubMed]
- Rakha E, Pajares MJ, Ilie M, et al. Stratification of resectable lung adenocarcinoma by molecular and pathological risk estimators. Eur J Cancer 2015;51:1897-903. [Crossref] [PubMed]
- Eguchi T, Kadota K, Chaft J, et al. Cell cycle progression score is a marker for five-year lung cancer-specific mortality risk in patients with resected stage I lung adenocarcinoma. Oncotarget 2016;7:35241-56. [Crossref] [PubMed]
- ClinicalTrials.gov. National Institutes of Health. Accessed 1st July 2017. Available online: https://clinicaltrials.gov/
- Kratz JR, Tham PT, Mulvihill MS, et al. Analytical validation of a practical molecular assay prognostic of survival in nonsquamous non-small cell lung cancer. Diagn Mol Pathol 2013;22:65-9. [Crossref] [PubMed]
- Woodard GA, Wang SX, Kratz JR, et al. Adjuvant Chemotherapy Guided by Molecular Profiling and Improved Outcomes in Early Stage, Non-Small-Cell Lung Cancer. Clin Lung Cancer 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Tsao MS, Le Teuff G, Shepherd FA, et al. PD-L1 protein expression assessed by immunohistochemistry is neither prognostic nor predictive of benefit from adjuvant chemotherapy in resected non-small cell lung cancer. Ann Oncol 2017;28:882-9. [PubMed]
- Horne ZD, Jack R, Gray ZT, et al. Increased levels of tumor-infiltrating lymphocytes are associated with improved recurrence-free survival in stage 1A non-small-cell lung cancer. J Surg Res 2011;171:1-5. [Crossref] [PubMed]
- Brambilla E, Le Teuff G, Marguet S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:1223-30. [Crossref] [PubMed]
- Bremnes RM, Busund LT, Kilvaer TL, et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:789-800. [Crossref] [PubMed]
- Romero Vielva L. Tumor lymphocytic infiltration in non-small cell lung cancer: the ultimate prognostic marker? Transl Lung Cancer Res 2016;5:370-2. [Crossref] [PubMed]
- Hu W, Yang Y, Zhang L, et al. Post surgery circulating free tumor DNA is a predictive biomarker for relapse of lung cancer. Cancer Med 2017;6:962-74. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Cheng J, Cuk K, Heil J, et al. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget 2017. [Epub ahead of print].
- Xiong Y, Huang BY, Yin JY. Pharmacogenomics of platinum-based chemotherapy in non-small cell lung cancer: focusing on DNA repair systems. Med Oncol 2017;34:48. [Crossref] [PubMed]
- Wallerek S, Sorensen JB. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur Respir Rev 2015;24:340-55. [Crossref] [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [Crossref] [PubMed]
- Bepler G, Olaussen KA, Vataire AL, et al. ERCC1 and RRM1 in the international adjuvant lung trial by automated quantitative in situ analysis. Am J Pathol 2011;178:69-78. [Crossref] [PubMed]
- Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101-10. [Crossref] [PubMed]
- Alden RS, Mandrekar SJ, Oxnard GR. Designing a definitive trial for adjuvant targeted therapy in genotype defined lung cancer: the ALCHEMIST trials. Chin Clin Oncol 2015;4:37. [PubMed]
- Goss GD, O'Callaghan C, Lorimer I, et al. Gefitinib versus placebo in completely resected non-small-cell lung cancer: results of the NCIC CTG BR19 study. J Clin Oncol 2013;31:3320-6. [Crossref] [PubMed]
- Kelly K, Altorki NK, Eberhardt WE, et al. A randomized, double-blind phase 3 trial of adjuvant erlotinib (E) versus placebo (P) following complete tumor resection with or without adjuvant chemotherapy in patients (pts) with stage IB-IIIA EGFR positive (IHC/FISH) non-small cell lung cancer (NSCLC): RADIANT results. J Clin Oncol 2014;32:7501.
- Pennell NA, Neal JW, Chaft JE, et al. SELECT: A multicenter phase II trial of adjuvant erlotinib in resected early-stage EGFR mutation-positive NSCLC. J Clin Oncol 2014;32:7514.
- Wu YL, Zhong W, Wang Q, et al. Gefitinib (G) versus vinorelbine+cisplatin (VP) as adjuvant treatment in stage II-IIIA (N1-N2) non-small-cell lung cancer (NSCLC) with EGFR-activating mutation (ADJUVANT): A randomized, Phase III trial (CTONG 1104). J Clin Oncol 2017;35:8500.
- Dagogo-Jack I, Santini F, Eng J, et al. Retrospective analysis of clinical outcomes of early stage ALK-Positive (ALK+) Non-Small Cell Lung Cancer (NSCLC). J Clin Oncol 2017;35:8536.
- Gerber DE, Oxnard GR, Govindan R. ALCHEMIST: Bringing genomic discovery and targeted therapies to early-stage lung cancer. Clin Pharmacol Ther 2015;97:447-50. [Crossref] [PubMed]
- O'Brien ME, Hasan B, Dafni U, et al. EORTC-ETOP randomized, phase 3 trial with anti-PD-1 monoclonal antibody pembrolizumab versus placebo for patients with early stage non-small cell lung cancer (NSCLC) after resection and standard adjuvant chemotherapy: PEARLS (NCT02504372). J Clin Oncol 2016. [Epub ahead of print].
- Dy GK. The role of PD-1/PD-L1 Inhibitors in the Adjuvant Setting for NSCLC. In: ASCO Daily News. 2016. Accessed 1st July 2017. Available online: https://am.asco.org/daily-news/role-pd-1pd-l1-inhibitors-adjuvant-setting-nsclc