Relevance of genetic alterations in squamous and small cell lung cancer
Squamous cell carcinomas of the lung
Squamous cell lung cancers (SQCLCs) account for 20–30% of NSCLCs and are the second most common kind of lung cancer diagnosed world-wide (1). Despite the relatively high incidence of this disease, there are in practice no targeted therapy options for patients with SQCLC, a situation that stands in stark contrast to the myriad options available for patients diagnosed with lung adenocarcinomas. This barrenness belies the substantial amount of work performed by multiple groups to better define the genomic abnormalities that are present in the disease. What it does reflect in part, is the representational disconnect that marks the existing models of this disease, which in turn exposes the need for additional experimental resources and a deeper understanding of how, and whether, these genomic abnormalities are functionally important.
A comprehensive review of the length and breadth of the molecular characteristics of SQCLCs provides an insufficient explanation as to why efforts at finding targeted therapies have largely failed, and can indeed be misleading when viewed strictly within the single-gene, single-target conceptual framework that has yielded success in the past. Rather, a review of the pre-clinical and clinical work on a handful of putative oncogenic targets provides the best assessment of where we currently stand in the development of targeted therapeutics for this disease. These events are, coincidentally, the most common genomic alterations found in SQCLCs, and include FGFR1 amplification (~20%), PI3K aberrations (~30–50%), and G1/S checkpoint alterations (~25%). What follows is a review of the pre-clinical and clinical data reported for each, with a focus on how these genotype-phenotype correlations inform ways to bridge the gap between the promise and reality of targeted options for patients with SQCLC.
FGFR1 amplification
FGFR1 is a transmembrane receptor tyrosine kinase that regulates embryonal development, cell proliferation, differentiation, and angiogenesis. The FGFR super-family consists of four receptors, including FGFR1, FGFR2, FGFR3 and FGFR4, and 22 FGF mitogenic and hormonal ligands. Ligand binding to the receptor, which is specific both to receptor isotype and organ location, causes receptor dimerization and activation of downstream signaling via the PI3K/AKT and RAS/RAF/MAPK pathways.
Alterations in FGFR1-4 were seen in 27% of early stage SQCLC tumor samples reported by TCGA, with FGFR1 mutations or amplification present in 18% (2). Other studies have shown amplification frequencies of between 13–22%, with variation by assay type and demographic group analyzed (3-6). Its impact on prognosis is unclear, with one study in an Asian population suggesting worse survival and another showing no impact on survival in a Caucasian population.
The first pre-clinical data to demonstrate the oncogenic potential of FGFR1 amplification in SQCLC was reported by Weiss and colleagues in 2011, who also showed that a subset of lung cancer cell lines and xenograft models that harbored this event were sensitive to treatment with the FGFR inhibitor PD173074 (4). Pharmacologic sensitivity in this setting was also reported in a number of FGFR1 amplified SQCLC patient derived xenografts in response to the FGFR inhibitor AZD4547 by Zhang and colleagues (7).
These data led to the inclusion of FGFR1 amplified SQCLCs in early phase clinical trials of the FGFR inhibitors AZD4547, BGJ398, dovitinib, and JNJ-42756493. The overall response rate in each of these biomarker-selected trials was modest and disappointing, ranging from 8–15% (8-11).
There are two general explanations as to why this might be the case. The first hinges on the presence of bypass pathways that confer resistance to drug inhibition. Malchers and colleagues, for example, have shown that high co-expression of Myc can sensitize FGFR1 amplified SQCLC to pharmacologic inhibition (12). In addition, genomic co-alterations with potential modifiers of response are also common, with frequent overlap with upstream PI3K alterations and G1/S checkpoint alterations (2,6). It is important to note, however, that the biologic impact of the latter has not yet been characterized.
The second explanation is definitional, that FGFR1 amplification, at least as defined by the assays in these trials, is not in fact a reliable proxy of downstream expression and thus activation. Wynes and colleagues suggested that this might be the case in their analysis of cell lines harboring varying degrees of FGFR1 copy number, mRNA, and protein expression, demonstrating that a correlation between DNA polysomy and expression was not uniform, rendering FGFR1 copy number as the least robust predictive biomarker of response to the drug ponatinib (13). Comprehensive molecular analysis of SQCLC patients treated with the FGFR inhibitor AZD4547 by Paik and colleagues supports this to be the case in patient samples, where receptor amplification was found to variably lead to increased mRNA and protein expression (14). In addition, expression analysis of the 8p11 amplicon showed striking differences between the expression patterns found in positive control FGFR1 amplified cell lung cancer cell lines that are known to be sensitive to AZD4547 and those from patient biopsies. While the cell line positive controls exhibited high expression of nearly all genes on the 8p11 amplicon, patient samples showed differential expression of these genes in patterns suggesting that FGFR1 was not central to the amplification event and, in most, not highly expressed relative to other genes.
These data suggest, in aggregate, that FGFR1 amplification is, in most patients, not a primary oncogenic event that can be targeted. While a subset of these patients might have cancers driven by FGFR1 amplification, the predictive biomarker that identifies these patients remains unclear.
PI3K pathway
The PI3K pathway, involved in cell survival, metabolism, motility, and angiogenesis, is commonly altered across many cancers. Oncogenic alterations can occur at multiple levels. In sum, these alterations are more common in SQCLCs than in lung adenocarcinomas, suggesting an increased dependence on this pathway (15-17). Upwards of 50% of tumors harbored a somatic alteration in one of the components of the PI3K/AKT pathway in the TCGA SQCLC dataset (2). The mutation rate for PIK3CA in SQCLC ranges from 3.6–6.5% (16,18), with PIK3CA amplification occurring in roughly 40% by FISH (17) and PCR (19) respectively. Jin and colleagues found a PTEN mutation rate of 10% in SQCLCs compared to 1.7% in lung adenocarcinomas (20). Soria and colleagues reported loss of PTEN expression by IHC and PTEN methylation in 24% and 35% of NSCLC respectively (21).
Functionally, conditional inactivation of PTEN and LKB1 in a transgenic murine model generates SQCLCs with complete penetrance (22). In vitro data has shown that cell lines harboring alterations in this pathway, including PIK3CA mutation or amplification and PTEN loss, confers sensitivity to pharmacologic inhibition (23). Retrospective analyses suggest that advanced SQCLC patients whose tumors harbor PI3K aberrations have a poorer prognosis, with an increased propensity to develop metastatic disease (6).
Despite these data, a phase 2 trial of the pan-PI3K inhibitor buparlisib (BKM120) in SQCLC patients with PI3K aberrations was negative (24). The trial enrolled 30 SQCLC patients in total, all of whom had tumors harboring an alteration in the PI3K pathway (21% PIK3CA mutation, 28% PTEN mutation, 62% PTEN IHC negative with varying degrees of overlap between categories). A single unconfirmed partial response was reported, yielding an ORR of 3%, in a patient whose tumor exhibited PTEN loss by IHC. The study did not meet its pre-specified PFS primary endpoint, and closed due to futility. A phase 2 trial of taselisib (GDC0032) under S1400 (NCT02154490) initially randomized PIK3CA mutant patients to taselisib 4 mg oral daily versus single agent docetaxel, but was amended to a single arm phase II trial of taselisib. Interim results of the first 20 patients were recently reported by Wade et al. and failed to meet the primary endpoint with an objective response rate of only 5% (95% CI, 1–24%) and a median PFS of 2.7 months (95% CI, 1.8–3.4 months) (25).
In the absence of more comprehensive sequencing data and/or pharmacodynamic correlates, it is difficult to tease apart the reasons for the limited efficacy seen in this biomarker-selected group of patients. That some or many of these PI3K alterations are passenger events is a distinct possibility, and one that has been raised in the past for patients with lung adenocarcinomas. Overlap is not uncommon with other putative drivers, similar to what has been observed with PIK3CA co-mutations in lung adenocarcinomas (26). Ultimately, modeling will be required to further understand mechanisms of resistance. The development of the Pten/Lkb1 null transgenic SQCLC mouse model should assist in these efforts.
Finally, it is worth noting that the consequence of somatic alterations such as PTEN loss is not limited in scope to oncogenic effects within the cancer cell. The Pten/Lkb1 null transgenic mouse model generates highly inflamed tumors that express PD-L1, which joins other data that suggests that PI3K pathway aberrations play a role in immunosurveillance (22,27,28).
CCND1 amplification, CDK4 amplification, CDKN2A loss
Cellular division is a highly regulated process marked by three general stages: quiescence (G0); DNA synthesis (G1, S); and mitosis (G2, M). Dysregulation of the cell cycle leading to uncontrolled division can occur through inactivation of suppressors such as RB, P53 and p16 and upregulation of activators such as the cyclin/cyclin dependent kinase (CDK) complexes. Cyclins D1-3, which bind to CDKs 2, 4, and 6, are important early regulators under the control of mitogenic signaling. These complexes phosphorylate the tumor suppressor RB, which triggers dissociation of RB from the E2F transcription factors and moves the cell from G1 to the S phase (29).
Of the G1/S checkpoint regulators, CCND1, which encodes cyclin D1, is amplified in 13% of SQCLC cases; CDK6 is amplified in 4%; and CDKN2A, which encodes the tumor suppressor p16 that inhibits CDK4 and 6, is mutated or homozygously deleted in 45% of tumors (2). There are, however, limited data to suggest that at least single agent inhibition of this pathway is effective in models of lung cancer. As an example, the CDK 4/6 inhibitor abemaciclib (LY2835219), was shown to have clear anti-tumor efficacy in breast and colorectal cancer models with and attendant decrease in RB phosphorylation (30). Modest growth inhibition was seen in response to abemaciclib monotherapy in a Calu-6 xenograft model, however, which is perhaps not surprising given the inability of abemaciclib to decrease RB phosphorylation in this model. A similar lack of anti-tumor efficacy was seen with palbociclib in H125 and H23 lung adenocarcinoma xenografts (31).
More recently, results from a phase 1 study of abemaciclib in NSCLC patients were reported as part of a multi-cancer basket trial (32). Sixty-eight patients with advanced NSCLC were treated, 29 of whom were KRAS mutant and 6 of had SQCLC. Two partial responses were seen, one in a KRAS mutant lung cancer patient and another in an SQCLC patient harboring CDKN2A loss. Molecular data was not available for 4 of the 6 patients with SQCLC. Of note one SQCLC patient with an increase in tumor size as best response also bore a tumor with CDKN2A loss.
The effect of other co-alterations as modifiers of response to CDK4/6 inhibition or, conversely, the role of G1/S checkpoint alterations as modifiers of response to other targeted agents is unknown and will require an analysis in a series of patients who undergo broader molecular testing. To this end, the results from the S1400 phase 2 study of palbociclib in advanced SQCLC patients whose tumors are positive for CCND1 amplification, CDK6 amplification, or CDKN2A deletion/mutation should be illuminating, as patients will have all undergone next-generation sequencing for a broad panel of oncogenes and tumor suppressor (S1400, NCT02154490).
DDR2 mutations
The discoidin domain receptor (DDR) is a plasma membrane receptor tyrosine kinase that regulates cell adhesion, proliferation, and extracellular remodelling upon binding to its endogenous ligand, type 1 collagen (33,34). Hammerman and colleagues reported that a subset of mutations in DDR2 are activating and that these mutations are sensitive to dasatinib, which has activity against DDR1 and DDR2 (35). While DDR2 mutations occur in about 3% of SQCLCs (2), they appear sporadically across the gene, and it is unlikely that all are functionally significant. There have been two case reports of SQCLC patients with DDR2 S768R mutations who have responded to dasatinib (35,36). This particular mutation appears to be rare, however, making any attempt at prospective study of this nearly impossible.
Future directions
Although these initial efforts at defining targets and matched therapies are now largely behind us, the way forward is not entirely clear. While the FDA has approved the non-targeted use of targeted therapies in this disease (necitumumab and afatinib), this is unsatisfying, subjecting most patients to ineffective therapies at best and needless toxicity at worst. At a minimum, attempts must be made at defining better biomarkers even as we expeditiously move away from those that have failed attempts at clinical validation. This is admittedly easier said than done, largely because our existing pre-clinical models do not reflect the diversity of oncogenic changes seen in our patients’ tumors. This can be surmounted by the generation of patient-derived xenografts, for example, if done in a systematic fashion.
And while there are other targets are of interest left to explore, such as NFE2L2 and KEAP1 mutations, the fate of targeted therapeutics for patients with SQCLCs might very well lie with combinations of inhibitors. The way forward with this is in theory clear, requiring a careful assessment of inhibitors using in vitro and in vivo models and cautious dose-finding studies. Both erlotinib + trametinib and dabrafenib + trametinib have been safely combined, the latter with success, providing precedence for this approach.
To this end, S1400 Lung-MAP (NCT02154490): a randomized phase II/III umbrella, biomarker-targeted, study of second-line therapy in patients with recurrent stage IV SQCLCs sponsored by the Southwest Oncology Group in collaboration with the National Cancer Institute is an ongoing protocol with a total estimated enrollment of 10,000 patients and an estimated study completion date of April 2025. This “Master Protocol” matches patients with a known biomarker or genomic alteration onto sub-studies of targeted therapies. The study is dynamic, and so can pivot in other directions as new data emerge, such as those discussed previously. Indeed, the plasticity of this study design serves as one of its greatest strengths, with the ability to open new cohorts to pursue novel targeted agents as they become available. Table 1 summarizes the protocol and current treatment arms.

Full table
Small cell carcinomas of the lung
In contrast to NSCLC, small cell lung cancer (SCLC) accounts for only 13% of all lung cancers (37,38). Despite the relatively low incidence of SCLC, it remains the 6th most common cause of cancer related death worldwide, attesting to the aggressive natural history and lack of effective therapies for this devastating disease (39). SCLC is a high grade neuroendocrine carcinoma with a rapid doubling time, early development of systemic metastases and a dismal 5-year overall survival of 1–5% (40). SCLC is a disease seen almost exclusively in patients with a significant smoking history, leading to a high mutational burden of carcinogen-induced somatic mutations including near universal bi-allelic loss of tumor suppressor genes TP53 and RB1 (41). First line systemic therapy for extensive stage disease consists of combination chemotherapy with a platinum and etoposide with response rates approaching 60–70% (42-44). Initial responses to systemic therapy are tantalizing, however, most patients suffer rapid and chemotherapy resistant disease relapse and treatment options have remained unchanged for the past three decades. Recent comprehensive genome, epigenome, and proteome analysis of SCLC has led to significant progress in uncovering aspects of the biology of the disease; defining new therapeutic strategies and offering renewed hope for patients. Table 2 summarizes ongoing clinical trials utilizing targeted therapies in SCLC.
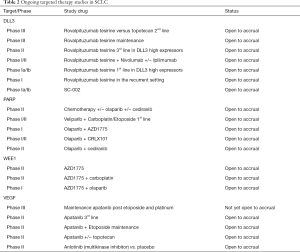
Full table
The genetic mutational landscape is complex and varied, however, the most common genetic alterations include functional inactivation of tumor suppressor genes TP53 and RB1, copy number gains in MYC family members, enzymes involved in chromatin remodeling, receptor tyrosine kinases and their downstream effector, and NOTCH family members (45-48). SCLC has one of the highest somatic mutation rates of 7.4 non-synonymous mutations per million base pairs (47), leading to an increased neoantigen load and a potential window of opportunity for therapeutic intervention with immunotherapy (49).
TP53 and RB1
TP53 and RB1, both tumor suppressors, are universally inactivated in SCLC (47). Rarely, SCLC can occur in the setting of retained or wild-type RB1. Chromothripsis, the phenomenon by which multiple clustered chromosomal arrangements occur in a single event, in chromosomes 3 and 11 can lead to overexpression of cyclin D1 and deregulation of Rb1, leading to the phenotypic presentation of SCLC (47). TP53, referred to as the “guardian of the genome,” plays a critical role in DNA repair and cell cycle arrest at the G1/S interphase, and is mutated in nearly half of all human cancers (50). Accordingly, TP53 deficiency leads to defective cell cycle checkpoint arrest at G1/S, impairs DNA damage response and contributes to replication stress (51-53). RB1, similar to TP53, encodes a protein (pRb) which binds to transcription factors of the E2F family and is responsible for cell cycle arrest and prevention of G1/S transition (54). In SCLC, RB1 and TP53 loss together result in markedly defective G1/S cell cycle checkpoint capacity and increased dependency on the G2/M checkpoint for adequate DNA repair and cell survival. In the context of TP53 and RB1 deficiency, rational targeting of the G2/M checkpoint may exploit this tumor specific vulnerability (53).
WEE1 inhibition
WEE1 is an important gatekeeper of G2 arrest and is a checkpoint tyrosine kinase that phosphorylates and inhibits cyclin-dependent kinases 1 (CDK1) and 2 (CDK2) (55). In the preclinical setting, the combination of WEE1 inhibitors with any of several classes of DNA damaging agents has shown synergistic efficacy in TP53 deficient cancer cell lines (56-58). AZD1775, is a novel tyrosine kinase inhibitor of Wee1, and is currently under clinical investigation in a Phase II, single arm study as monotherapy in patients with recurrent SCLC (NCT02593019) and an ongoing phase I with a SCLC expansion cohort in combination with PARP inhibitor olaparib (NCT02511795).
EZH2 inhibition
Loss of RB1 in SCLC is also strongly associated with overexpression of enhancer of zeste homolog 2 (EZH2) (59,60) suggesting a potential role for EZH2 inhibition in the treatment of SCLC. EZH2 is not commonly mutated in SCLC but the level of EZH2 expression in SCLC is higher than in any tumor type included in the Cancer Genome Atlas (61,62). EZH2 expression is under the direct control of E2F transcription factors (63). E2F transcriptional activity is negatively regulated by RB1 (47); the nearly universal loss of functional RB1 in SCLC results in high-level E2F transcriptional activity, and consequent high-level EZH2 expression (59). These observations define a model in which EZH2 expression is primarily promoted by one of the pathognomonic genetic alterations of SCLC. Recent work using patient-derived xenografts links the upregulation of EZH2 with SLFN11 silencing as a frequent mechanism of acquired chemoresistance in SCLC and EZH2 inhibition was found to prevent SLFN11 silencing and maintain chemotherapeutic sensitivity, suggesting a potential combinatorial strategy to enhance efficacy of current standard therapies for this recalcitrant disease (64).
Notch pathway
The Notch pathway has been implicated in SCLC oncogenesis and multiple approaches targeting the pathway are currently underway (47,65). Direct Notch inhibitors such as MEDI0639, a human monoclonal antibody directed against delta like ligand 4 (DLL4) is believed to block angiogenesis by the formation of nonfunctional vasculature and inhibiting tumor initiating stem cells (66). A phase I study of MED10639 (NCT01577745) was reported in 2015 and preliminary evidence of antitumor activity was observed (67). OMP-59R5, a human monoclonal antibody targeting Notch 2/3 receptors has showed exciting preclinical activity (68). PINNACLE (NCT01859741), a phase Ib/II study of showed promise in combination with platinum and etoposide with an 81% response rate in previously untreated patients with ES-SCLC.
Delta-like ligand 3 (DLL3) plays a key role in neural development and is normally only expressed in the developing central nervous system (69,70). DLL3 is highly upregulated and aberrantly expressed on the cell-surface of high-grade neuroendocrine tumors including SCLC, leading to its potential as a therapeutic target (71). Rovalpituzumab teserine (Rova-T) is a novel, first-in-class, antibody-drug conjugate with high specificity for DLL3. Rova-T binds to DLL3, is internalized, and undergoes cleavage of a linker moiety, releasing a pyrrolobenzodiazepine dimer (PBD) cytotoxic payload, resulting in tumor-specific DNA damage and cell death (71). Rudin and colleagues recently reported the first-in human phase I trial of Rova-T in patients with recurrent metastatic SCLC and large cell neuroendocrine lung cancers (72). The study defined a recommended phase II dose and noted dose limiting toxicities including thrombocytopenia, liver test abnormalities, and serosal effusions. Of the evaluable patients, 17% (11/65) achieved a confirmed objective response and 54% (35/65) had stable disease. In patients with high DLL3 expression (protein expression in ≥50% of cells), the response rate was 39% (10/26). Most notably, among patients in whom tissue was available for protein analysis, responses were observed exclusively in those with high level DLL3 expression. Subsequent studies to further evaluate Rova-T agent are ongoing, including an open-label, multicenter, phase II trial evaluating the efficacy of Rova-T for third line and later treatment of patients with DLL3-positive SCLC (NCT02674568); Rova-T as frontline treatment of patients with high DLL3 expressing ES-SCLC (NCT02819999); Rova-T as maintenance therapy following first line platinum based chemotherapy (NCT03033511); and lastly Rova-T in combination with nivolumab and with or without ipilimumab in the second line setting (NCT03026166).
FGFR
Fibroblast growth factor receptor-1 FGFR1 is amplified in 6% of patients with SCLC (73). However, FGFR1 protein expression and mRNA levels, and not gene copy number, have been shown to predict FGFR TKI sensitivity in preclinical models (13). There are ongoing clinical trials assessing FGFR inhibition including a recently completed phase II study of lucitanib (74), a multi-kinase inhibitor of FGFR1-3, vascular endothelial growth factor receptor VEGFR1-3, and platelet derived growth factor receptor PDGFRα/β, (NCT02109016) that was completed in April of 2016 and has yet to report results. An earlier phase I/IIa basket study of lucitanib did not include patients with SCLC (75). Also ongoing is a single center study phase II biomarker driven study of ponatinib (NCT01935336) enrolling patients with ES and LS-SCLC.
MYC
MYC gene family members have known oncogenic potential and MYC amplification has been implicated in oncogenesis in SCLC. MYC is a transcriptional regulator of aurora kinase A and B, and in the absence of p53, has the potential to drive oncogenesis (76). Aurora kinase A is essential for centrosome function, spindle assembly, chromosome alignment, and mitotic entry (77). Knocking down the expression of Aurora A gene inhibits cell proliferation and induces G2/M phase arrest in human SCLC cells (78). Targeted drug screens reveal that SCLC with high MYC expression, and high NEUROD1 expression, are vulnerable to Aurora kinase inhibition, when combined with chemotherapy, strongly suppresses tumor progression (79). Alisertib, an investigational, orally available, selective Aurora A kinase inhibitor, has shown preclinical activity across multiple tumor types (80). A recently reported multicenter phase I/II study of alisertib defined a recommended phase II dose and schedule and noted dose limiting toxicities including neutropenia, leukopenia, and anemia. The phase II expansion cohort reported a single agent response rate of 21% (10/48) (81). A randomized phase II study of alisertib or placebo in combination with paclitaxel in SCLC patients in the second line setting recently reported a response rate of 22% in the experimental arm (20/89) and 18% (16/89) in the control arm (82). Further investigation is needed to better understand the utility of aurora kinase inhibitors in SCLC.
MYCL fusion
Several gene fusions have been identified, but virtually all are of unknown significance, including a recurrent in-frame RLF-MYCL1 gene fusion identified in 9% of primary SCLC samples and cell lines (83). RNAi targeting of MYCL1 mRNA in RLF-MYCL1 fusion positive human SCLC cells reduced cell proliferation suggesting that the tumor-specific RLF-MYCL1 fusion may be a novel oncogenic driver that is required for SCLC tumorigenesis. Future work will be required to validate the functional significance of this fusion product.
PARP inhibition
Byers and colleagues reported on the proteomic and transcriptomic analysis of human SCLC cell lines (62). SCLC showed significantly increased levels of apoptosis mediators, DNA repair proteins including poly-(ADP)-ribose polymerase (PARP) enzymes, and of EZH2. PARP1, an E2F1 co-activator, involved in DNA repair was highly expressed at both the mRNA and protein levels in SCLC, leading to the hypothesis and preclinical validation that PARP inhibition downregulated key DNA repair mechanisms leading to enhanced efficacy of chemotherapy and perhaps other DNA damaging therapies (62). Schlafen family member 11 (SLFN11) has been identified as a critical determinant of PARP inhibitor sensitivity in SCLC cell lines and patient-derived xenografts (84,85). SLFN11 is actively recruited to sites of DNA damage, inhibits HR and activates a replication stress response (85). High SLFN11 expression is associated with improved tumor response, progression free survival and overall survival in SCLC patients treated with temozolomide and the PARP inhibitor veliparib in a recent randomized phase II clinical trial (86). Further work to define the utility of SLFN11 expression as a predictive biomarker of PARP inhibitor therapy in SCLC will require validation in prospective trials.
Immunotherapy
SCLC has one of the highest somatic mutation rates of 7.4 non-synonymous mutations per million base pairs (47), leading to an increased neoantigen load and a potential window of opportunity for therapeutic intervention with immunotherapy (49). Escape from immune surveillance is a well-recognized feature of cancer (87) and the development of therapies to enhance anti-tumor immune response has led to exciting new treatment options. T-cell checkpoint blockade, targeting programmed death ligand-1 (PD-1) with or without concomitant inhibition of cytotoxic T-lymphocyte associated protein-4 (CTLA-4), has shown activity in SCLC (88). The high mutational burden seen in SCLC, resulting in a large number of potential tumor-specific antigenic determinants, may favor sensitivity to T-cell checkpoint inhibition. However, in stark contrast to other tumor types, SCLC rarely expresses PD-L1. In the only study fully reported to date, CheckMate032, PD-L1 expression did not appear to be predictive of immunotherapy response. Objective response rates ranged from 19% to 23% for the combination arms and translated into an encouraging one-year survival of 35–43%. These results have led to the incorporation of combination nivolumab and ipilimumab as a National Comprehensive Cancer Network treatment guideline recommendation in the second line setting.
Future directions
Comprehensive genome, proteome and transcriptome analysis of human SCLC has led to the development of novel targeted therapeutics currently under clinical investigation. However, many promising targeted agents have been studied with little improvement in the overall survival of our patients. Continued utilization and improvement of our in vitro and in vivo model systems that mirror human SCLC pathogenesis, with carcinogen induced mutations, is critical to our pathway forward. Models are needed to better understand mechanisms of our immunotherapeutic interventions and potential resistance. Equally important will be the study of normal neuroendocrine cell signaling and differentiation, including neuroendocrine cell plasticity and neuroendocrine to mesenchymal transition as well as the poorly understood small cell transformation seen infrequently in non-SCLC. As we continue to better understand the biology of SCLC, we have to stay true to ourselves and our patients in designing combination trials that are grounded in scientific reason and that focus on maintaining quality of life and minimize potential toxicity.
Acknowledgements
Funding: This work was supported by P30 CA008748 (to PK Paik), Celgene research funding (to PK Paik), T32 CA009207 (to JK Sabari).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Travis WD. Pathology of lung cancer. Clin Chest Med 2011;32:669-92. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. [Crossref] [PubMed]
- Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6:e20351. [Crossref] [PubMed]
- Kim Y, Hammerman PS, Kim J, et al. Integrative and Comparative Genomic Analysis of Lung Squamous Cell Carcinomas in East Asian Patients. J Clin Oncol 2014;32:121-8. [Crossref] [PubMed]
- Paik PK, Shen R, Won H, et al. Next-Generation Sequencing of Stage IV Squamous Cell Lung Cancers Reveals an Association of PI3K Aberrations and Evidence of Clonal Heterogeneity in Patients with Brain Metastases. Cancer Discov 2015;5:610-21. [Crossref] [PubMed]
- Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res 2012;18:6658-67. [Crossref] [PubMed]
- Paik P, Shen R, Ferry D, et al. A phase 1b open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers: Preliminary antitumor activity and pharmacodynamic data. J Clin Oncol 2014;32:abstr 8035.
- Nogova L, Sequist LV, Garcia JMP, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol 2017;35:157-65. [Crossref] [PubMed]
- Lim SH, Sun J-M, Choi Y-L, et al. Efficacy and Safety of Dovitinib in Pretreated Advanced Squamous Non-small Cell Lung Cancer with FGFR1 Amplification: A Single-arm, Phase II Study. Cancer 2016;122:3024-31. [Crossref] [PubMed]
- Tabernero J, Bahleda R, Dienstmann R, et al. Phase I Dose-Escalation Study of JNJ-42756493, an Oral Pan–Fibroblast Growth Factor Receptor Inhibitor, in Patients With Advanced Solid Tumors. J Clin Oncol 2015;33:3401-8. [Crossref] [PubMed]
- Malchers F, Dietlein F, Schottle J, et al. Cell-Autonomous and Non-Cell-Autonomous Mechanisms of Transformation by Amplified FGFR1 in Lung Cancer. Cancer Discov 2014;4:246-57. [Crossref] [PubMed]
- Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and Protein Expression, not Gene Copy Number, Predict FGFR TKI Sensitivity across All Lung Cancer Histologies. Clin Cancer Res 2014;20:3299-309. [Crossref] [PubMed]
- Paik PK, Shen R, Berger MF, et al. A Phase 1b Open Label Multicentre Study of AZD4547 in Patients with Advanced Squamous Cell Lung Cancers. Clin Cancer Res 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res 2012;18:1167-76. [Crossref] [PubMed]
- Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913-21. [Crossref] [PubMed]
- Okudela K, Suzuki M, Kageyama S, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int 2007;57:664-71. [Crossref] [PubMed]
- Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 2006;54:209-15. [Crossref] [PubMed]
- Ji M, Guan H, Gao C, et al. Highly frequent promoter methylation and PIK3CA amplification in non-small cell lung cancer (NSCLC). BMC Cancer 2011;11:147. [Crossref] [PubMed]
- Jin G, Kim MJ, Jeon HS, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer 2010;69:279-83. [Crossref] [PubMed]
- Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res 2002;8:1178-84. [PubMed]
- Xu C, Fillmore Christine M, Koyama S, et al. Loss of Lkb1 and Pten Leads to Lung Squamous Cell Carcinoma with Elevated PD-L1 Expression. Cancer Cell 2014;25:590-604. [Crossref] [PubMed]
- Spoerke JM, O'Brien C, Huw L, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res 2012;18:6771-83. [Crossref] [PubMed]
- Vansteenkiste JF, Canon JL, Braud FD, et al. Safety and Efficacy of Buparlisib (BKM120) in Patients with PI3K Pathway-Activated Non-Small Cell Lung Cancer: Results from the Phase II BASALT-1 Study. J Thorac Oncol 2015;10:1319-27. [Crossref] [PubMed]
- Wade JL, Langer CJ, Redman M, et al. A phase II study of GDC-0032 (taselisib) for previously treated PI3K positive patients with stage IV squamous cell lung cancer (SqNSCLC): LUNG-MAP sub-study SWOG S1400B. J Clin Oncol 2017;35:abstr 9054.
- Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma – rationale for comprehensive mutation profiling. Mol Cancer Ther 2012;11:485-91. [Crossref] [PubMed]
- Song M, Chen D, Lu B, et al. PTEN Loss Increases PD-L1 Protein Expression and Affects the Correlation between PD-L1 Expression and Clinical Parameters in Colorectal Cancer. PLoS One 2013;8:e65821. [Crossref] [PubMed]
- Lastwika KJ, Wilson W, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT–mTOR Pathway in Non–Small Cell Lung Cancer. Cancer Res 2016;76:227-38. [Crossref] [PubMed]
- Dickson MA, Schwartz GK. Development of cell-cycle inhibitors for cancer therapy. Curr Oncol 2009;16:36-43. [PubMed]
- Gelbert LM, Cai S, Lin X, et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: in-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Invest New Drugs 2014;32:825-37. [Crossref] [PubMed]
- Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3:1427-38. [PubMed]
- Patnaik A, Rosen LS, Tolaney SM, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non–Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov 2016;6:740-53. [Crossref] [PubMed]
- Ikeda K, Wang LH, Torres R, et al. Discoidin domain receptor 2 interacts with Src and Shc following its activation by type I collagen. J Biol Chem 2002;277:19206-12. [Crossref] [PubMed]
- Olaso E, Labrador JP, Wang L, et al. Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J Biol Chem 2002;277:3606-13. [Crossref] [PubMed]
- Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discov 2011;1:78-89. [Crossref] [PubMed]
- Pitini V, Arrigo C, Di Mirto C, et al. Response to dasatinib in a patient with SQCC of the lung harboring a discoid-receptor-2 and synchronous chronic myelogenous leukemia. Lung Cancer 2013;82:171-2. [Crossref] [PubMed]
- Govindan R, Page N, Morgensztern D, et al. Changing Epidemiology of Small-Cell Lung Cancer in the United States Over the Last 30 Years: Analysis of the Surveillance, Epidemiologic, and End Results Database. J Clin Oncol 2006;24:4539-44. [Crossref] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2016. Atlanta (GA): American Cancer Society, 2016.
- Nicholson AG, Chansky K, Crowley J, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the Revision of the Clinical and Pathologic Staging of Small Cell Lung Cancer in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:300-11.
- Brambilla E, Auerbach O, Kuschner M, et al. Small cell carcinoma. In: Travis WD, Brambilla E, Burke AP, editors. World Health Organization Classification of Tumors: Tumors of the Lung, Pleura, Thymus and Heart - Pathology and Genetics. Lyon, France: IARC, 2015.
- Varghese AM, Zakowski MF, Yu HA, et al. Small-Cell Lung Cancers in Patients Who Never Smoked Cigarettes. J Thorac Oncol 2014;9:892-6. [Crossref] [PubMed]
- Fukuoka M, Saijo N, Nishiwaki Y, et al. Randomized trial of cyclophosphamide, doxorubicin, and vincristine versus cisplatin and etoposide versus alternation of these regimens in small-cell lung cancer. J Natl Cancer Inst 1991;83:855-61. [Crossref] [PubMed]
- Roth BJ, Johnson DH, Einhorn LH, et al. Randomized study of cyclophosphamide, doxorubicin, and vincristine versus etoposide and cisplatin versus alternation of these two regimens in extensive small-cell lung cancer: a phase III trial of the Southeastern Cancer Study Group. J Clin Oncol 1992;10:282-91. [Crossref] [PubMed]
- Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus Cisplatin Compared with Etoposide plus Cisplatin for Extensive Small-Cell Lung Cancer. N Engl J Med 2002;346:85-91. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Dowlati A, Lipka MB, McColl K, et al. Clinical correlation of extensive-stage small-cell lung cancer genomics. Ann Oncol 2016;27:642-7. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323-31. [Crossref] [PubMed]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature 1991;351:453-6. [Crossref] [PubMed]
- Dobbelstein M, Sorensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov 2015;14:405-23. [Crossref] [PubMed]
- Goodrich DW, Wang NP, Qian Y-W, et al. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 1991;67:293-302. [Crossref] [PubMed]
- Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science 1992;257:1955-7. [Crossref] [PubMed]
- Hirai H, Iwasawa Y, Okada M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther 2009;8:2992-3000. [Crossref] [PubMed]
- Hirai H, Arai T, Okada M, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther 2010;9:514-22. [Crossref] [PubMed]
- Rajeshkumar NV, De Oliveira E, Ottenhof N, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res 2011;17:2799-806. [Crossref] [PubMed]
- Coe BP, Thu KL, Aviel-Ronen S, et al. Genomic Deregulation of the E2F/Rb Pathway Leads to Activation of the Oncogene EZH2 in Small Cell Lung Cancer. PloS One 2013;8:e71670. [Crossref] [PubMed]
- Hubaux R, Thu KL, Coe BP, et al. EZH2 Promotes E2F-Driven SCLC Tumorigenesis through Modulation of Apoptosis and Cell-Cycle Regulation. J Thorac Oncol 2013;8:1102-6. [Crossref] [PubMed]
- Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015;34:5869-78. [Crossref] [PubMed]
- Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798-811. [Crossref] [PubMed]
- Bracken AP, Pasini D, Capra M, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003;22:5323-35. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Takebe N, Nguyen D, Yang SX. Targeting Notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol Ther 2014;141:140-9. [Crossref] [PubMed]
- Jenkins DW, Ross S, Veldman-Jones M, et al. MEDI0639: A Novel Therapeutic Antibody Targeting Dll4 Modulates Endothelial Cell Function and Angiogenesis In Vivo. Mol Cancer Ther 2012;11:1650-60. [Crossref] [PubMed]
- Falchook GS, Dowlati A, Naing A, et al. Phase I study of MEDI0639 in patients with advanced solid tumors. ASCO 2015 Annual Meeting Abstract 3024. J Clin Oncol 2015;33.
- Yen WC, Fischer MM, Axelrod F, et al. Targeting Notch Signaling with a Notch2/Notch3 Antagonist (Tarextumab) Inhibits Tumor Growth and Decreases Tumor-Initiating Cell Frequency. Clin Cancer Res 2015;21:2084-95. [Crossref] [PubMed]
- Loomes KM, Stevens SA, O'Brien ML, et al. Dll3 and Notch1 genetic interactions model axial segmental and craniofacial malformations of human birth defects. Developmental Dynamics 2007;236:2943-51. [Crossref] [PubMed]
- Ladi E, Nichols JT, Ge W, et al. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol 2005;170:983-92. [Crossref] [PubMed]
- Saunders LR, Bankovich AJ, Anderson WC, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 2015;7:302ra136-302ra136.
- Rudin CM, Pietanza MC, Bauer TM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol 2017;18:42-51. [Crossref] [PubMed]
- Schultheis AM, Bos M, Schmitz K, et al. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol 2014;27:214-21. [Crossref] [PubMed]
- Guffanti F, Chilà R, Bello E, et al. In Vitro and In Vivo Activity of Lucitanib in FGFR1/2 Amplified or Mutated Cancer Models. Neoplasia 2017;19:35-42. [Crossref] [PubMed]
- Soria JC, DeBraud F, Bahleda R, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol 2014;25:2244-51. [Crossref] [PubMed]
- Brockmann M, Poon E, Berry T, et al. Small Molecule Inhibitors of Aurora-A Induce Proteasomal Degradation of N-Myc in Childhood Neuroblastoma. Cancer Cell 2013;24:75-89. [Crossref] [PubMed]
- Nikonova AS, Astsaturov I, Serebriiskii IG, et al. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci 2013;70:661-87. [Crossref] [PubMed]
- Lu Y, Liu Y, Jiang J, et al. Knocking down the expression of Aurora A gene inhibited cell proliferation and induced G2/M phase arrest in human small cell lung cancer cells. Oncol Rep 2014;32:243-9. [Crossref] [PubMed]
- Mollaoglu G, Guthrie MR, Böhm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- Manfredi MG, Ecsedy JA, Chakravarty A, et al. Characterization of Alisertib (MLN8237), an Investigational Small-Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin Cancer Res 2011;17:7614-24. [Crossref] [PubMed]
- Melichar B, Adenis A, Lockhart AC, et al. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol 2015;16:395-405. [Crossref] [PubMed]
- Owonikoko T, Nackaerts K, Csoszi T, et al. OA05.05 Randomized Phase 2 Study: Alisertib (MLN8237) or Placebo + Paclitaxel as Second-Line Therapy for Small-Cell Lung Cancer (SCLC). J Thorac Oncol 2017;12:S261-S262. [Crossref]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Lok BH, Gardner EE, Schneeberger VE, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res 2017;23:523-35. [Crossref] [PubMed]
- Murai J, Feng Y, Yu GK, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 2016;7:76534-50. [PubMed]
- Byers LA, Krug L, Waqar S, et al. MA11.07 Improved Small Cell Lung Cancer (SCLC) Response Rates with Veliparib and Temozolomide: Results from a Phase II Trial. J Thorac Oncol 2017;12:S406-S407. [Crossref]
- Schreiber RD, Old LJ, Smyth MJ. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. [Crossref] [PubMed]


