Expanded molecular interrogation for potential actionable targets in non-squamous non-small cell lung cancer
Introduction
The adoption of potent and specific kinase inhibitors against epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangement has necessitated significant change in the diagnostic and clinical algorithms in the management of non-small cell lung cancer (NSCLC) (1). It is now routine to mandate a good quality tissue biopsy for thorough histological review (rather than cytology specimens), routine genomic testing for non-squamous non-small cell lung cancer (ns-NSCLC), prior to making treatment recommendations (2). The burden of proof supporting this approach is most substantial for EGFR and ALK, where phase III trials in biomarker-selected patients, have incontrovertibly confirmed higher efficacy of targeted agents against platinum-based chemotherapy (1).
The striking success with targeted therapies has led to significant enthusiasm for the identification of additional molecularly-defined subsets. To this end, comprehensive large-scale sequencing studies have provided an unabridged list of frequently recurring genomic events that may represent potential targets (3), although there are several limitations. First, with each tumor harboring multiple non-synonymous events, not all genetic alterations may be equally functional and as a consequence druggable. Second, the genomic context of these alterations has become increasingly relevant, where pan-cancer analysis has suggested that alterations such as PIK3CA often occur as subclonal events, consistent with the variable experience observed in the clinic to date in PI3K targeting (4,5). Similarly, some alterations may be involved in tumor initiation and have a lesser contribution to the on-going progression malignant phenotype. Third, depending on the relevance of the target to human physiology, and the specificity of the drug, there may not be a sufficient therapeutic index to effect anti-tumor activity (6). Lastly, not all NSCLC cases are accounted for by known recurrent somatic events, reflecting the need to examine other non-genomic mechanisms such as epigenetic and transcriptomic changes to delineate the entire spectrum of tumor vulnerabilities.
Nevertheless, the establishment of EGFR, ALK and ROS1 as bona fide oncogene-addicted NSCLC have set the benchmark for future credentialing of actionable targets (2). Additional targets such as BRAF, MET, RET and HER2 have shown early promise (7), with several currently undergoing clinical evaluation in early phase trials. In this review, we will discuss the emerging data on the genomic alterations in non-squamous NSCLC, some of the novel epigenetic targets in development, and the anticipated challenges in the expanding scope for molecular interrogation and defining new standards of care for rare molecular subsets.
Determinants of cancer traits and scope of actionable targets
Anticancer therapies aim to subvert mission-critical mechanisms of tumorigenesis and cell survival. Within a tumor ecosystem, this is dictated by multiple layers of regulation, starting with intercellular differences in genomic alterations, epigenetic and transcriptomic changes, as well as clonal diversity, tumor microenvironment and the immune landscape. The treatment for NSCLC has been revolutionized due to the biomarker-driven treatment paradigm, exemplified by EGFR mutations and ALK rearrangements. Having elucidated the genomic landscape of NSCLC through large-scale sequencing studies (8-10), we now have a slew of uncommon yet equally actionable targets e.g., MET exon 14 skipping mutations, BRAF alterations (7). With new drug discovery technologies, such as fragment-based design and ultra-high throughput screens, coupled with enhanced molecular profiling of patients, the number of patients who could potentially benefit from new highly specific and optimized investigational drugs are set to increase. Similarly, tumor mutational burden and signatures can provide complementary information, such as those relating to biomarkers for immune checkpoint inhibitors and potential etiologies to cancer (11,12).
While genomics has been widely adopted and remains the cornerstone of our efforts in precision oncology, it is important to explore a broader repertoire of potential actionable targets—one area of high interest is epigenetic targets. Epigenetics is defined as heritable changes in gene expression without alteration of the DNA sequence, broadly encompassing DNA methylation, histone modification and non-coding RNA (13). Chromatin structure has been shown to influence the mutation rates in cancer cells (14-16). The associated changes in chromatin accessibility and modifications are thought to be a major feature in shaping the mutational landscape between different cell of origins (16). At present, majority of drug development into epigenetic targets has been focused on DNA methylation and histone modifications. Of note, signalling pathways can also be influenced by chromatin structure and dynamics (17), highlighting the complex interplay between oncogenic signalling and the cancer epigenome. Unlike genetic mutations, epigenetic aberrations are potentially reversible, providing an attractive approach to modulate the effects of multiple signalling pathways (18).
The next sections will describe a range of promising genomic and epigenetic targets in development and the importance of broad comprehensive molecular profiling.
Emerging actionable genomic targets in NSCLC
BRAF mutations
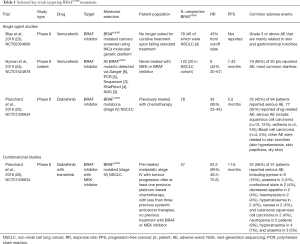
BRAF acts as a kinase that connects the RAS guanosine triphosphate to the proteins found within the mitogen-activated protein kinase (MAPK) pathway (19). A BRAF mutation promotes tumorigenesis by activating the proliferative pathway and MAPK2 and MAPK3 (20). Although this proto-oncogene is most commonly found in melanoma patients, BRAF mutations are still found amongst 1–3% of lung adenocarcinoma patients, about half of which harbor the V600E mutation, where valine is substituted for glutamic acid in exon 15 (21). BRAF mutations have been reported in smokers and never smokers, suggesting that enrichment molecular screening strategies are probably inadequate (21,22). Past studies have shown these patients are less responsive to platinum-based chemotherapy and have poor clinical outcomes (22), underscoring the importance of identifying such patients early on in the disease course. The latest clinical trials have shown promise and summarised below (Table 1).
Full table
Single agent basket trials have generally seen overall response rates of 42–43% (23,24). An ACSE Phase II trial of vemurafenib (NCT02304809) in advanced NSCLC and other BRAFV600E cancer patients demonstrated a 43% overall response rate (ORR) for NSCLC patients (23). Another similar study (NCT01524978) explored the tumor agnostic concept, where BRAFV600E mutant tumors regardless of tumor type were treated with vemurafenib. Notably, response rates were 42% in the NSCLC cohort and progression-free survival (PFS) 7.43 months (24). Subsequently, two phase II trials involving dabrafenib monotherapy and dabrafenib-trametinib combination were conducted in parallel (NCT01336634), examining the concept of vertical blockade. The combination of dabrafenib and trametinib achieved an overall response rate of 63.2% (26) compared to a 33.3% ORR for patients that only received dabrafenib (25). Of note, reported adverse events for either the monotherapy or the combinational therapy included skin-related toxicities and most strikingly the development of cutaneous squamous cell carcinoma and basal cell carcinoma. Notably, combinational therapy reduced the incidence of cutaneous squamous cell carcinoma to only 2 of 57 patients (4%) instead of 10 of 84 patients (12%) affected and none of the patients developed basal cell carcinoma in the combination group (25,26). The encouraging clinical results and decreased side effects led to granting of accelerated approval by the Food and Drug Administration (FDA), where the combination approach is the current accepted standard for advanced staged BRAFV600E NSCLC patients (27).
In contrast to BRAFV600E, non-V600 mutations, which account for the remaining half of BRAF mutations in NSCLC, are insensitive to currently approved BRAF inhibitors. Combinatorial therapy with dabrafenib (RAF inhibitor) and trametinib (MEK inhibitor) has demonstrated anti-tumor activity in NSCLC cell lines harbouring non-V600 BRAF mutations (28). The efficacy of trametinib in non-V600 mutated NSCLC is currently being addressed in the NCI-MATCH trial (29).
MET amplification and MET exon 14 skipping
MET protein is located on the cell surface and is the tyrosine kinase receptor which binds to its ligand, hepatocyte growth factor (HGF). The binding leads to in MET dimerization, resulting in a cascade effect that ultimately leads to cell proliferation (30). While previous high profile phase III trial failures e.g. with the monoclonal antibody against MET, onartuzumab (31) were largely attributed to inadequate patient selection criteria, more robust biomarkers have since been developed. There are currently two types of MET abnormalities that show promise in the clinic—MET copy number gain and MET exon 14 skipping—with a 2–4% and 3–4% respective frequency within lung adenocarcinoma patients (32).
One key impetus in elucidating the therapeutic tractability of MET alterations in NSCLC, is the fact that crizotinib, approved for ALK and ROS1 alterations, has also demonstrated potent activity against c-MET (33,34). Thus, with the clinical availability of a potent MET inhibitor, there was a need to delineate the predictive value of MET alterations. Here, one of the key challenges is the definition of thresholds of MET copy number gain, and more recently MET exon 14 skipping mutations.
MET copy number gain was first established as a mechanism of resistance to EGFR TKI, where it has been described in up to 20% of patients (35,36). Its role in the treatment naïve setting has also been investigated using copy number cutoffs (37). Patients with high copy number are reported to be an indicator of a more aggressive disease (38). Complicating the interpretation of these cutoffs, is the fact that MET copy number gain, can occur in the context of focal amplification, polysomy (likely a reflection of ploidy) and even with other genomic alterations e.g., EGFR, KRAS in up to 56% of cases using a 5 copy threshold (37). Thus while MET fluorescent in situ hybridization (FISH) has been largely adopted to pre-select patients for trials, it is important to note that concurrent screening with broader next generation sequencing to exclude other drivers or immunohistochemistry (IHC) to ascertain protein expression may be complementary and allow further enrichment for potential c-MET addicted tumors (39).
The initial experience in targeting MET copy number gains was in the context of PROFILE 1001 (NCT00585185), where an extension cohort enrolled 14 patients with low, intermediate, or high copy number ratios. In this small cohort of patients, the ORR was 0%, 17% and 67% for the low, intermediate and high MET groups respectively when treated with crizotinib (40). Additional studies have since attempted to better delineate the patient population driven by MET amplification. Excluding patients with other co-existing oncogenes and using a ratio of >1.8, around 4.8% of patients remain MET-amplified (35). Ongoing studies of cMET tyrosine kinase inhibitors, e.g., INC280 Capmatinib (NCT02750215) and MGCD 265 Glesatinib (NCT02544633) are directed at defining clinically relevant thresholds that predict for efficacy to cMET inhibitors.
There is also accumulating data on the therapeutic tractability of cMET exon 14 skipping mutations. From a mechanistic viewpoint, these mutations result in a splicing defect resulting in exon 14 skipping, resulting in loss of a ubiquitination site, and hence constitutive over-expression of c-MET (32). The PROFILE 1001 study included a further cohort of patients with exon 14 skipping mutations that reported an ORR of 44% and median PFS of 7.36 months to crizotinib. Certain guidelines have also incorporated the use of a MET inhibitors in patients with MET activated tumors (40).
While exon 14 skipping mutations appear to be a defined molecular subset, the implications of the varying copy numbers in patients with MET amplification is still not well understood. The importance of stratification has been highlighted through preliminary data where imposing strict definitions of MET amplification—patients with a ratio of >5 yielded an ORR of 67% compared to 0% in patients with ratios of <2 (40). Trials of MET inhibitors enrolling patients with specific copy number gain thresholds will further our understanding of targeting the MET axis, although they also pose significant logistical and regulatory challenges. Here, the major concern is that stringency of enrolment criteria can directly impact on size of target population, which in turn can influence trial feasibilities and anticipated efficacy. In this instance, the availability of relevant historical controls in patients harbouring a similar molecular profile are useful to benchmark outcomes in the setting of a single-arm study.
HER2 mutations/overexpression/amplifications
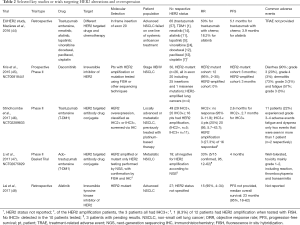
Mutation of the human epidermal growth factor 2 (HER2) is found in 1–4% of NSCLC patients, and in 6% of EGFR, ALK and KRAS negative individuals, with a preponderance for never smokers and women (41). The most common mutation of HER2 in NSCLC is an exon 20 in-frame insertion (A775_G776insYVMA), leading to constitutive kinase activity and downstream activation of AKT and MEK pathways (42). In addition to insertion at the kinase domain, point mutation of the extracellular domain (S310F) has also been reported (43). To date there have been limited number of clinical trials specifically targeting this molecular subset (Table 2). Earlier trials showed that the monoclonal antibody against HER2, trastuzumab in combination with chemotherapy failed to demonstrate any clinical benefit in NSCLC patients (49,50). A retrospective review of 65 HER2 mutation positive patients, representing 1.7% of 3,800 patients screened, for the first time reported an objective response rate of 50% with trastuzumab monotherapy and various combinations with chemotherapy (51). In an extended pan European cohort study (EUHer2), patients who underwent HER2-based treatment (trastuzumab, neratinib, afatinib and lapatinib), demonstrated an objective response rate of 50.9% and a PFS of 4.8 months (44). While these studies on combination chemotherapy and HER2-based treatment showed promising response rates, the actual contribution of HER2 targeting and relative efficacy of each individual anti-HER2 agent are difficult to ascertain from currently available data. Another prospective phase II study of dacomitinib (NCT00818441), an irreversible inhibitor of HER2, EGFR, and HER4 was reported in 30 patients, of which 26 harbored HER2 mutations (25 insertions and 1 missense mutation); 3 of the 26 patients responded [RR 12% (95% CI, 2–30%)]) with no partial responses seen in the amplified cases (45). A recent retrospective study of HER2 mutant NSCLC treated with afatinib also showed activity of 15% in 4 out of 27 patients (48). Interestingly, there may be mutation specific drug sensitivities for HER2 exon 20 insertions, such as those that result in insertions of amino acids either before or after the C-helix, resulting in a change in conformational transitions between active and inactive states, and sensitivity to irreversible EGFR TKI (52).
Full table
More recently, two phase II studies highlighted the potential of trastuzumab emtansine (T-DM1) in HER2 mutant NSCLC (46,47). In the first study by Stinchcombe et al. (NCT02289833), 49 patients with HER2 overexpression (IHC2+ and above) received T-DM1. Patients with IHC2+ demonstrated an ORR of 0% and PFS of 2.6 months, while those with IHC3+ had an ORR of 20% and PFS of 2.7 months (46). Li et al.’s basket trial (NCT02675829) in 18 patients showed an ORR of 33% and PFS of 4 months (47). It is important to highlight that both studies employed different selection criteria for trial enrolment. The first study enrolled patients who were IHC2+ or IHC3+ on HER2 IHC, and showed that the latter group were more responsive to TDM-1, with 4 out of 20 patients exhibiting a partial response (20%), while none of the IHC2+ patients responded. Furthermore, amongst the 49 patients, 16 were detected to have HER2 amplification (by ISH), 5 were IHC2+ and 11 were IHC3+ patients. Among the IHC3+ patients who also had HER2 amplification, ORR was 27.3%—raising the possibility that IHC3+ patients with HER2 amplification may be more responsive to TDM-1 (46). The second study primarily evaluated HER2 mutations in the cohort, with no evidence of HER2 amplification by NGS or HER3+ expression in those with tissue available (47). Overall, the clinical experience to date supports the role of both kinase inhibitors and HER2 targeting antibodies in HER2 mutated or HER2 expressing NSCLC.
RET rearrangements
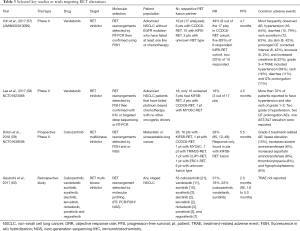
RET alterations account for 1–2% of NSCLC, and is especially prevalent in young, non-smokers with poorly differentiated tumours (53,54). While multi-kinase inhibitors potently targeting RET such as vandetanib (ZD 6474) and cabozantinib (XL184) have been approved by the FDA to treat advanced metastatic medullary thyroid cancer, its efficacy in NSCLC is currently still under investigation (55,56). Two phase II studies in Japanese and Korean population were recently reported. The LURET study (UMIN000010095) was a nationwide effort conducted in Japan, which investigated the role of vandetanib in 17 advanced NSCLC patients (57). The results were encouraging, nine (47%) of the 17 patients responded, while the median PFS was 4.7 months. Similarly, a Korean phase II study (NCT01823068) also investigated the vandetanib, demonstrating a response rate of 17%, in which three of the 18 patients responded, with a PFS of 4.6 months (58). A third study evaluated the role of cabozantinib in 25 patients (NCT01639508), showing an ORR of 28% and PFS of 5.5 months (59). Recently, an international global registry reported a modest role for current RET-targeting multi-kinase inhibitors in 53 RET rearranged NSCLC (which included cabozantinib, vandetanib, sunitinib, sorafenib, alectinib, lenvatinib, nintedanib, ponatinib and regorafenib). In this study, the PFS was reported to be 2.3 months and overall survival 6.8 months (60), underscoring the need for more specific and potent RET inhibitors. Although the small sample size of these studies no doubt contributed to the range of outcomes observed, differences in pre-screening strategies can also impact on responses. Notably, the Japanese study enrolled patients from a national molecular screening program (LC-SCRUM) where patients underwent RT-PCR and were eligible only if confirmed by FISH (57), while the Korean study screened EGFR wild type, ALK non-rearranged patients with break-apart FISH, and the results were subsequently confirmed using one of the following tests, IHC, RT-PCR or targeted deep sequencing (58). In the Japanese study, the adjusted ORR following vandetanib was 83% among 6 patients with CCDC6-RET and 20% among 10 patients with KIFB-RET fusion (57). Similarly the Korean study reported mixed clinical outcome with RET targeting, which was dependent on the underlying RET fusion partner (58), highlighting the potential implications of different RET fusions, as well as the importance of employing high precision biomarkers in clinical trials. A summary of these trials is presented in Table 3.
Full table
NTRK rearrangements
Another emerging therapeutic target in NSCLC is the NTRK fusion protein. The NTRK genes (NTRK1, 2 and 3) encode for a family of receptor tyrosine kinase known as tropomysin receptor kinase (TrkA, TrkB and TrkC), which plays a crucial role in neuronal development (61). Genetic rearrangements of the NTRK family have been reported in about 3% of NSCLC (62). Importantly, selective tyrosine kinase inhibitors against NTRK fusion proteins have demonstrated promising in vitro and in vivo anti-tumor activity in NSCLC (62-64).
Epigenetic targets in development for NSCLC
While there have been significant advances in the various genomic targets described above, the role of epigenetic targets has been less well-defined. Nevertheless, given the important role in cancer development and drug resistance, there have been numerous forays into understanding the role and therapeutic tractability of DNA methylation and histone modifiers.
DNA methylation
Genome wide DNA methylation studies have yielded differentially methylated genes in NSCLC compared to paired normal controls (65). Importantly, DNA methylation status of selected genes confers prognostic and predictive information on patient management. For example, the concurrent hypermethylation of four genes, CDKN2A, CDH13, RASSF1A and APC in early stage NSCLC correlated with early recurrence and death (66). Moreover, DNA methylation profile in a select panel of genes and hypomethylation of ERBB2 predict sensitivity to erlotinib (67), while methylation of IGFBP-3 is associated with resistance to cisplatin in NSCLC (68).
DNA methylation is mediated by a family of DNA methyltransferase (DNMT), namely DNMT1, DNMT3A and DNMT3B (69). 5-azacitidine and decitabine are two DNMT inhibitors that have demonstrated treatment efficacy in the realm of haematopoietic tumours (70). While clinical trials utilizing decitabine as monotherapy have yielded disappointing results (71,72), combination of DNMT inhibitor 5-azacitidine and histone deacetylase (HDAC) inhibitor entinostat showed objective response in patients with advanced NSCLC (73).
Histone modification and chromatin organization
Post-translational modification of histones regulates chromatin structure and transcriptional activity. A number of histone post-translational modifications have been described to date, including methylation, acetylation, phosphorylation and ubiquitylation (74). These histone marks define functional regions of the epigenome. For example, H3K4me3 marks active promoters. Active enhancers are enriched for H3K27ac, while H3K27me3 is a repressive mark on enhancers. Early work showed that global levels of histone modifications by IHC in clinical samples of NSCLC confer prognostic information (75-77). Importantly, these histone marks are reversible and are tightly regulated by epigenetic regulators. Four classes of epigenetic regulators have been described, namely writers, erasers, readers and movers (74). Writers (histone acetyltransferase, histone methyl transferase) are enzymes that add a specific post-translational modification to histone, while erasers (HDAC, histone demethylase) do the opposite. Readers such as bromodomain and extra terminal domain (BET) family proteins, recognize specific histone marks and direct the appropriate transcriptional response. Movers (SWI/SNF complex) are ATP-dependent chromatin remodelers which reposition nucleosomes along the genome. Several large scale genomic studies in NSCLC have identified somatic mutations in epigenetic regulators, raising the notion of epigenetic deregulation as the eleventh hallmark of cancer (8).
EZH2, a subunit of the polycomb repressive complex 2 (PRC2) is a histone lysine methyltransferase that mediates H3K27 trimethylation to silence target genes (78). EZH2 is overexpressed in NSCLC and is associated with poor prognosis (79,80). Similarly overexpression of HDACs has been observed in NSCLC (81). It has been shown that EZH2 interacts with HDAC to repress transcription (82). Combined inhibition of EZH2 and HDAC has a synergistic antiproliferative effect in NSCLC cell lines (83). Inactivating mutations of MLL2, another histone lysine methyltransferase has also been reported in NSCLC (84).
BET family proteins (BRD2, BRD3, BRD4 and BRDT) are readers that recognize acetylated lysine residues in histones, which play an important role in transcription control (85). Translocation of BRD4, a well characterized member of the BET family, to the NUT gene with the resulting BRD4-NUT fusion protein defines a recently described entity known as NUT midline carcinoma (NMC). NMC is a poorly differentiated squamous cell carcinoma that occurs in the head and neck as well as the thoracic cavity (86). It shows marked sensitivity to BET targeting (87). Notably, BRD4 is also overexpressed in NSCLC, and correlates with poor prognosis in NSCLC patients (88). BET targeting has been shown to be effective in acute myeloid leukemia and multiple myeloma (89). A novel BET inhibitor, OTX015 exhibits in vitro anti-tumor activity against NSCLC cell lines harboring different oncogenic mutations (90).
SMARCA4/BRG1 is an ATP-dependent catalytic subunit of the SWI/SNF chromatin remodelling complex, best known for its role in malignant rhabdoid tumors. Loss of SMARC4 has been reported in 5% to 20% of NSCLC, leading to epigenetic silencing of downstream genes, independent of DNA methylation (91,92). Interestingly, SMARCA4 deficient tumors are typically EGFR wild type and TTF-1 negative (93), suggesting that SMARCA4 loss could be a bone fide oncogenic driver event. Somatic inactivation of other members of the SWI/SNF complex, ARID1A and ARID2 has also been reported in NSCLC (94).
To summarize, epigenetic deregulation is involved, at least in part in the pathogenesis of NSCLC. Importantly selective inhibitors against a number of these epigenetic regulators are currently in various stages of trial and their potential role either as single agents or in combination with other targeted therapies are currently under investigation. We anticipate that evaluation for aberrations in these regulators would be critical in selecting patients for the appropriate epigenetic therapy.
Expanding the biomarker repertoire for actionable targets
Although traditional platforms e.g., IHC, FISH, Sanger sequencing, have been adequate in the era of single biomarkers for a limited number of drugs, the lack of scalability and precision for emerging targets is a disadvantage (95). Further, given the low frequency of some alterations, tissue attrition remains a challenge if testing for multiple separate assays especially with needle biopsies frequently employed (96). Given the diverse targets that are rapidly approaching the clinic, there is a need to continue development of biomarker platforms capable for broad unbiased tumor profiling to capture the relevant actionable alterations.
One significant advance is the introduction of next generation sequencing in the clinic (97). At a genomic level, we have already described a range of genetic lesions that need to be tested concurrently—including somatic mutations, chromosomal rearrangements, amplification and loss of function. Somatic mutations are often responsible for the activation of the EGFR oncogene, KRAS, BRAF, while chromosomal rearrangement drives both ALK and ROS1 (3,39). Amplification and deletions can also lead to gain of function and loss of function with associated expression changes of certain genes (98). As alluded to in the previous sections there may also be genetic changes that overlap e.g., MET exon 14 skipping mutations and amplification, or HER2 mutations and amplifications, as well as co-alterations. Eliciting the function in the setting of complex genetic profiles where there may be two or more druggable alterations, remains a key clinical priority. Further, as we improve our understanding on epigenetic mechanisms that drive tumorigenesis and progression, it might become increasingly feasible to develop biomarker assays that can be adopted in the clinic. Given the diverse mechanisms of epigenetic regulation, such advances will allow improved selection and stratification for specific epigenetic approaches, as well as facilitate combinatorial approaches. Moving ahead, functional evaluation, and unbiased RNA-seq complemented by high throughput epigenetic assays may be implemented.
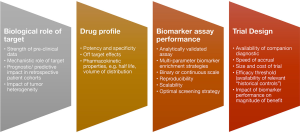
As discussed amply in the earlier sections, the molecular screening methodology and understanding the genomic context of any biomarker can play a critical role in the eventual trial design. Some relevant factors for consideration are summarized in Figure 1. Ultimately the ability to calibrate the precision of biomarkers that exist on a continuous scale e.g., MET copy gain and PD-L1, will have significant implications on trial design, feasibility and outcomes. Given the ability to define small molecular subsets, and the complexity and dynamic nature of drug resistance, it is conceivable that there will be a diminishing role for large randomized trials. Furthermore, the expanding ability to interrogate individual tumors across multiple-dimensions, will place increasing emphasis on comprehensive biomarker compendiums that enable precise stratification of individual patient cohorts according to likelihood of benefit.
Conclusions
Given the exciting developments with targeted therapies beyond EGFR and ALK, various guidelines now emphasize the importance of broader profiling. For example, the National comprehensive cancer network (NCCN) guidelines in 2017, not only is screening for EGFR and ALK a part of the standard treatment for NSCLC, but testing for ROS1, RET, BRAF, HER2 and MET alterations are also recommended. Through broader profiling panels, we will continue to learn more about a range of genetic abnormalities that will provide a blueprint to understanding the life histories of individual cancers and potential therapeutic vulnerabilities. Other targets such as KRAS and PIK3CA will be more readily identified and present enhanced opportunities for clinical trials to better understand the mechanisms of response and resistance to targeted approaches. With further advances in technologies and declining sequencing costs, it is anticipated that the scale and breadth of tumor profiling will enable implementation of highly nuanced stratification approaches that ultimately aim to deliver cost-effective precision cancer therapies.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Thomas A, Liu SV, Subramaniam DS, et al. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 2015;12:511-26. [Crossref] [PubMed]
- Villaruz LC, Burns TF, Ramfidis VS, et al. Personalizing therapy in advanced non-small cell lung cancer. Semin Respir Crit Care Med 2013;34:822-36. [Crossref] [PubMed]
- Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156-64. [PubMed]
- McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 2015;7:283ra54. [Crossref] [PubMed]
- Vansteenkiste JF, Canon JL, Braud FD, et al. Safety and Efficacy of Buparlisib (BKM120) in Patients with PI3K Pathway-Activated Non-Small Cell Lung Cancer: Results from the Phase II BASALT-1 Study. J Thorac Oncol 2015;10:1319-27. [Crossref] [PubMed]
- Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med 2011;32:703-40. [Crossref] [PubMed]
- Oxnard GR, Binder A, Jänne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol 2013;31:1097-104. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Holliday R. Epigenetics: a historical overview. Epigenetics 2006;1:76-80. [Crossref] [PubMed]
- Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 2012;488:504-7. [Crossref] [PubMed]
- Makova KD, Hardison RC. The effects of chromatin organization on variation in mutation rates in the genome. Nat Rev Genet 2015;16:213-23. [Crossref] [PubMed]
- Polak P, Karlić R, Koren A, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 2015;518:360-4. [Crossref] [PubMed]
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 2013;14:211-24. [Crossref]
- Liu SV, Fabbri M, Gitlitz BJ, et al. Epigenetic therapy in lung cancer. Front Oncol 2013;3:135. [Crossref] [PubMed]
- Luk PP, Yu B, Ng CC, et al. BRAF mutations in non-small cell lung cancer. Transl Lung Cancer Res 2015;4:142-8. [PubMed]
- Beeram M, Patnaik A, Rowinsky EK. Raf: a strategic target for therapeutic development against cancer. J Clin Oncol 2005;23:6771-90. [Crossref] [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Blay JY, Mazieres J, Perol D, et al. Vemurafenib (VM) in non-melanoma V600 and non-V600 BRAF mutated cancers: first results of the ACSE trial. Ann Oncol 2016;27:55PD.
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- FDA. FDA grants regular approval to dabrafenib and trametinib combination for metastatic NSCLC with BRAF V600E mutation. 2017. Available online: https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm564331.htm
- Noeparast A, Teugels E, Giron P, et al. Non-V600 BRAF mutations recurrently found in lung cancer predict sensitivity to the combination of Trametinib and Dabrafenib. Oncotarget 2017;8:60094-108.
- NCI. NCI-MATCH Trial (Molecular Analysis for Therapy Choice). Available online: https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Spigel DR, Edelman MJ, O'Byrne K, et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung. J Clin Oncol 2017;35:412-20. [Crossref] [PubMed]
- Sacco JJ, Clague MJ. Dysregulation of the Met pathway in non-small cell lung cancer: implications for drug targeting and resistance. Transl Lung Cancer Res 2015;4:242-52. [PubMed]
- Zou HY, Li Q, Lee JH, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res 2007;67:4408-17. [Crossref] [PubMed]
- Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 2011;6:942-6. [Crossref] [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Noonan SA, Berry L, Lu X, et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. J Thorac Oncol 2016;11:1293-304. [Crossref] [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 2009;27:1667-74. [Crossref] [PubMed]
- Khoo C, Rogers TM, Fellowes A, et al. Molecular methods for somatic mutation testing in lung adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res 2015;4:126-41. [PubMed]
- Camidge DR, Ou S-HI, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:abstr 8001.
- Peters S, Zimmermann S. Targeted therapy in NSCLC driven by HER2 insertions. Transl Lung Cancer Res 2014;3:84-8. [PubMed]
- Wang SE, Narasanna A, Perez-Torres M, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 2006;10:25-38. [Crossref] [PubMed]
- Greulich H, Kaplan B, Mertins P, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A 2012;109:14476-81. [Crossref] [PubMed]
- Mazières J, Barlesi F, Filleron T, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol 2016;27:281-6. [Crossref] [PubMed]
- Kris MG, Camidge DR, Giaccone G, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol 2015;26:1421-7. [Crossref] [PubMed]
- Stinchcombe T, Stahel RA, Bubendorf L, et al. Efficacy, safety, and biomarker results of trastuzumab emtansine (T-DM1) in patients (pts) with previously treated HER2-overexpressing locally advanced or metastatic non-small cell lung cancer (mNSCLC). J Clin Oncol 2017;35:abstr 8509.
- Li BT, Shen R, Buonocore D, et al. Ado-trastuzumab emtansine in patients with HER2 mutant lung cancers: Results from a phase II basket trial. J Clin Oncol 2017;35:abstr 8510.
- Lai W-CV, Lebas L, Milia J, et al. Afatinib in patients with metastatic HER2-mutant lung cancers: An international multicenter study. Journal of Clinical Oncology 2017;35:abstr 9071.
- Gatzemeier U, Groth G, Butts C, et al. Randomized phase II trial of gemcitabine-cisplatin with or without trastuzumab in HER2-positive non-small-cell lung cancer. Ann Oncol 2004;15:19-27. [Crossref] [PubMed]
- Lara PN, Laptalo L, Longmate J, et al. Trastuzumab plus docetaxel in HER2/neu-positive non-small-cell lung cancer: a California Cancer Consortium screening and phase II trial. Clin Lung Cancer 2004;5:231-6. [Crossref] [PubMed]
- Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. [Crossref] [PubMed]
- Kosaka T, Tanizaki J, Paranal RM, et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res 2017;77:2712-21. [Crossref] [PubMed]
- Chao BH, Briesewitz R, Villalona-Calero MA. RET fusion genes in non-small-cell lung cancer. J Clin Oncol 2012;30:4439-41. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012;30:4352-9. [Crossref] [PubMed]
- Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 2014;14:173-86. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]
- Lee SH, Lee JK, Ahn MJ, et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: a phase II clinical trial. Ann Oncol 2017;28:292-7. [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Gautschi O, Milia J, Filleron T, et al. Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J Clin Oncol 2017;35:1403-10. [Crossref] [PubMed]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 2003;72:609-42. [Crossref] [PubMed]
- Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469-72. [Crossref] [PubMed]
- Tatematsu T, Sasaki H, Shimizu S, et al. Investigation of neurotrophic tyrosine kinase receptor 1 fusions and neurotrophic tyrosine kinase receptor family expression in non-small-cell lung cancer and sensitivity to AZD7451 in vitro. Mol Clin Oncol 2014;2:725-30. [Crossref] [PubMed]
- Farago AF, Le LP, Zheng Z, et al. Durable Clinical Response to Entrectinib in NTRK1-Rearranged Non-Small Cell Lung Cancer. J Thorac Oncol 2015;10:1670-4. [Crossref] [PubMed]
- Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012;22:1197-211. [Crossref] [PubMed]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118-28. [Crossref] [PubMed]
- Walter K, Holcomb T, Januario T, et al. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clin Cancer Res 2012;18:2360-73. [Crossref] [PubMed]
- Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010;29:1681-90. [Crossref] [PubMed]
- Kim GD, Ni J, Kelesoglu N, et al. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J 2002;21:4183-95. [Crossref] [PubMed]
- Estey EH. Epigenetics in clinical practice: the examples of azacitidine and decitabine in myelodysplasia and acute myeloid leukemia. Leukemia 2013;27:1803-12. [Crossref] [PubMed]
- Momparler RL, Bouffard DY, Momparler LF, et al. Pilot phase I-II study on 5-aza-2'-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anticancer Drugs 1997;8:358-68. [Crossref] [PubMed]
- Schrump DS, Fischette MR, Nguyen DM, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res 2006;12:5777-85. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 2015;16:178-89. [Crossref] [PubMed]
- Barlési F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol 2007;25:4358-64. [Crossref] [PubMed]
- Van Den Broeck A, Brambilla E, Moro-Sibilot D, et al. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 2008;14:7237-45. [Crossref] [PubMed]
- Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 2009;174:1619-28. [Crossref] [PubMed]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006;6:846-56. [Crossref] [PubMed]
- Huqun Ishikawa R. Enhancer of zeste homolog 2 is a novel prognostic biomarker in nonsmall cell lung cancer. Cancer 2012;118:1599-606. [Crossref] [PubMed]
- Behrens C, Solis LM, Lin H, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res 2013;19:6556-65. [Crossref] [PubMed]
- Sasaki H, Moriyama S, Nakashima Y, et al. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer 2004;46:171-8. [Crossref] [PubMed]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 1999;23:474-8. [Crossref] [PubMed]
- Takashina T, Kinoshita I, Kikuchi J, et al. Combined inhibition of EZH2 and histone deacetylases as a potential epigenetic therapy for non-small-cell lung cancer cells. Cancer Sci 2016;107:955-62. [Crossref] [PubMed]
- Yin S, Yang J, Lin B, et al. Exome sequencing identifies frequent mutation of MLL2 in non-small cell lung carcinoma from Chinese patients. Sci Rep 2014;4:6036. [Crossref] [PubMed]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 2007;26:5521-7. [Crossref] [PubMed]
- French CA, Miyoshi I, Kubonishi I, et al. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res 2003;63:304-7. [PubMed]
- Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067-73. [Crossref] [PubMed]
- Liao YF, Wu YB, Long X, et al. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget 2016;7:9491-500. [Crossref] [PubMed]
- Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem Sci 2015;40:468-79. [Crossref] [PubMed]
- Riveiro ME, Astorgues-Xerri L, Vazquez R, et al. OTX015 (MK-8628), a novel BET inhibitor, exhibits antitumor activity in non-small cell and small cell lung cancer models harboring different oncogenic mutations. Oncotarget 2016;7:84675-87. [PubMed]
- Orvis T, Hepperla A, Walter V, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res 2014;74:6486-98. [Crossref] [PubMed]
- Song S, Walter V, Karaca M, et al. Gene silencing associated with SWI/SNF complex loss during NSCLC development. Mol Cancer Res 2014;12:560-70. [Crossref] [PubMed]
- Matsubara D, Kishaba Y, Ishikawa S, et al. Lung cancer with loss of BRG1/BRM, shows epithelial mesenchymal transition phenotype and distinct histologic and genetic features. Cancer Sci 2013;104:266-73. [Crossref] [PubMed]
- Manceau G, Letouzé E, Guichard C, et al. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. Int J Cancer 2013;132:2217-21. [Crossref] [PubMed]
- Wu YC, Chang IC, Wang CL, et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS One 2013;8:e70839. [Crossref] [PubMed]
- Sequist LV, Engelman JA, Lynch TJ. Toward noninvasive genomic screening of lung cancer patients. J Clin Oncol 2009;27:2589-91. [Crossref] [PubMed]
- Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev 2011;32:177-95. [PubMed]
- Fong KM, Sekido Y, Minna JD. Molecular pathogenesis of lung cancer. J Thorac Cardiovasc Surg 1999;118:1136-52. [Crossref] [PubMed]