Emerging uses of circulating tumor DNA in advanced stage non-small cell lung cancer
Introduction
Lung cancer remains the leading cause of cancer-related death in the United States (1). The majority of patients with non-small cell lung cancer (NSCLC) are diagnosed with metastatic disease, which is generally fatal (1,2). Recent research has revealed that patients with metastatic NSCLC constitute a heterogeneous group. For a minority of patients with advanced NSCLC whose tumors harbor specific genetic aberrations detected by next generation sequencing and other molecular analyses, administration of oral tyrosine kinase inhibitors (TKIs) can result in improved clinical outcomes (3-5). As a result, epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1) directed therapies are now recommended for first-line treatment of advanced NSCLC patients harboring appropriate genetic abnormalities (6). The expanding number of somatic genomic targets and recent availability of second- and third-generation TKIs specific to certain resistance mutations have prompted reflex molecular profiling of tumor tissue to become commonplace both at diagnosis and at disease progression in order to characterize these oncogenic drivers and mechanisms of resistance and help guide appropriate treatment selection.
Tissue biopsies have long been the gold standard for determining the genetic profile of a patient’s cancer, but tissue biopsies are often difficult to obtain safely (7). Furthermore, genetic testing of tumor tissues can take several weeks, ultimately delaying initiation therapy whether or not a genetic aberration is identified. Although TKIs are initially very effective in most patients whose tumors harbor a genetic aberration, the majority of patients will eventually develop resistance to these agents within a year of their initiation (8-10). The mechanism of resistance is in many cases unknown and is a matter of intensive research. Some resistance mechanisms, such as the T790M acquired resistance mutation in EGFR, are better characterized and can specifically direct future treatments with drugs such as osimertinib (11,12). Many patients, however, have less common or unknown resistance mechanisms that cannot be directly targeted with FDA approved therapies (9,10,13). Repeat molecular analysis at the time of progression is therefore essential to identify the mechanism of resistance and potentially new treatment approaches.
Tissue biopsies are challenging to obtain, sometimes associated with adverse events, and expensive. Clonal heterogeneity, whereby different clones may harbor distinct resistance mechanisms (14,15), adds further complexity; it is simply not feasible to biopsy every tumor in a given patient. Consequently, there is growing interest in analyzing tumor material and other tumor biomarkers in various bodily fluids, so-called “liquid biopsies”, as a way to more easily and, if necessary, serially detect molecular alterations during a patient’s treatment course. In addition to being easier to obtain than a tissue biopsy, liquid biopsies offer the hope of providing a more complete, integrated picture of a patient’s cancer compared with conventional tissue biopsy of a single tumor site since bodily fluids may contain tumor material released from multiple disease sites. Analysis of cancer cell derived DNA from circulating tumor DNA (ctDNA) enables real-time molecular monitoring of cancer and demonstrates great promise toward realizing personalized cancer care in NSCLC.
ctDNA consists of short, double-stranded DNA fragments that are shed into the blood stream by tumor cells undergoing apoptosis or necrosis (16). ctDNA has a relatively short half-life (approximately 2 hours) and often represents a small fraction (<1.0%) of the total cell-free DNA (cfDNA) circulating in the bloodstream. It is distinguished from normal cfDNA by the identification of somatic mutations present in the cancer genome (17). Highly sensitive genotyping assays such as digital polymerase chain reactions (PCR) (18); beads/emulsion/amplification/magnetics (BEAMing) (19), and next-generation sequencing (NGS) technologies (20) have enabled the accurate detection of rare mutant variants present in body fluids. Several studies have demonstrated that ctDNA can be readily detected in patients with NSCLC, with improved sensitivity in patients with advanced stage disease and those with a higher burden of disease (21,22). ctDNA analysis allows clinicians to non-invasively interrogate tumor specific molecular alterations without the need for a traditional tissue biopsy. ctDNA technology can be utilized for identifying actionable mutations at diagnosis, tracking genomic evolution and the development of resistance mutations to targeted therapies, and monitoring response to therapy.
Detecting mutations in ctDNA
Limited panels
The initial panels developed to characterize a patient’s tumor through liquid biopsies used digital PCR to detect hotspot mutations in the most clinically relevant genes. Sacher et al. looked prospectively at the feasibility and concordance of digital PCR to detect EGFR exon 19 deletion, L858R, T790M and KRAS G12X in the peripheral blood of patients with NSCLC compared to tissue biopsy. Plasma testing had an excellent sensitivity, with a high concordance with tissue biopsy. The authors also noted a significant advantage of plasma testing over tissue biopsy—the test turnaround time for digital PCR was a median 3 days. This compared favorably to tissue biopsy, which had a median turnaround time of 12–27 days (22).
Next generation sequencing
Although limited panels using PCR are fast, they are unable to detect clinically important rearrangements such as those occurring in the ALK, RET and ROS1 genes. In addition, they limit the detection of less common resistance mutations, as genetic pathways less commonly associated with resistance may not be in a limited panel. Targeted NGS of ctDNA can detect single nucleotide variants, fusions and insertions/deletions in a more comprehensive fashion. Targeted NGS of ctDNA has been shown to provide a high degree of accuracy across multiple tumor types (23,24). A study by Paweletz et al. investigated a NGS ctDNA panel in 48 NSCLC patients and found a sensitivity of 77% across 62 known driver and resistance mutations (25). In a recent study done by our group, we found that ctDNA NGS testing in a population enriched for patients likely to have useful reporter mutations was technically successful in 102 patients, while tissue testing was technically successful in 50 patients. The most common reasons for unsuccessful tissue analysis were inaccessible tissue and inadequate DNA in the sample. ctDNA analysis was able to detect 50 driver mutations, 12 resistance mutations, and mutations in 22 additional genes that could serve as potential therapeutic targets. The concordance between ctDNA NGS testing and tissue testing of the most common mutation detected, EGFR, was 79% (26).
In our study, ctDNA NGS testing was able to detect resistance mutations in eight patients who developed progressive disease while on targeted therapy, but who were not able to have additional tissue sequencing (26). FDA-approved targeted therapies were identified for 32% of the patients who had ctDNA testing. This proportion likely understates the utility of such an approach, however, as 55% of patients had a targetable genetic aberration, including both patients with FDA approved targeted therapies and those genetic aberrations for which published data may justify off-label use. When one includes all genetic aberrations currently being evaluated in ongoing clinical trials, up to 70% of tested patients had a genetic aberration that could be targeted. Considering the heterogeneous and diverse mechanisms of resistance to targeted therapies, the theoretical benefit of a NGS approach in this setting becomes clear; by casting a wider net, we can identify targets that may be useful in clinical care.
Clinical factors may impact the sensitivity of ctDNA in NSCLC. Two separate studies have shown that ctDNA was more likely to be detected among patients with squamous cell carcinoma than adenocarcinoma (27,28). This is thought to be due to a higher rate of tumor necrosis and passive DNA release among patients with squamous cell carcinoma. A high Ki67 proliferative index as well as pre-operative 18F-fluoro-deoxyglucose (FDG) avidity has also been associated with a higher rate of ctDNA detection (28).
Concordance of plasma, urine, and tissue testing
Testing for ctDNA can be done accurately in both urine and plasma. This has been shown in multiple studies looking at the sensitivity of ctDNA in the urine and plasma to detect T790M mutations in patients with EGFR mutations that are developing resistance to a 1st-line TKI.
In a subset analysis of a clinical trial of rociletinib, Wakelee et al. evaluated the concordance between tissue, plasma and urine testing for T790M mutation among 242 patients with NSCLC harboring an EGFR mutation. Among the 189 patients positive for T790M by plasma, 175 were also positive on tissue biopsy (81.5% concordance between plasma and tissue testing). There were similar results in the group that had ctDNA testing of the urine, with 83.8% concordance between urine and tissue testing. Regardless of how the T790M mutation was detected the overall response and duration of response for treatment with rociletinib were similar (29).
In a series of patients with EGFR mutant NSCLC being treated with osimertinib, discordant results were evaluated further. Patients who were plasma negative and tissue positive for T790M had an ORR of 69% and progression free survival (PFS) of 16.5 months, whereas patients negative for T790M in both the plasma and tissue had inferior outcomes (ORR, 25%; PFS, 2.8 months). Patients with plasma positive and tissue negative tumors had an intermediate outcome, with an ORR of 28% and PFS of 4.3 months. The relatively high sensitivity of plasma testing (30% false negative rate) implies that a significant portion of patients can avoid tissue biopsy. It remains important to emphasize, however, that patients who were plasma negative still could benefit from osimertinib therapy if tissue biopsy was positive. As such, tissue biopsies remain a vital component of our diagnostic paradigm (30).
Discordant results remain a major clinical quandary—should a patient receive the targeted therapy detected by one test if the other test indicates a lack of potential benefit? While we have emerging data for patients with EGFR mutations suggesting that there is clinical benefit to act on the positive test result regardless of its source, this problem potentially affects all targeted therapies. It is already fairly evident that test concordance according to DNA source will not be uniform. The cause of tissue/ctDNA discordance remains unclear. One potential mechanism may be increased sensitivity of ctDNA assays, as compared to tissue assays. In a study by our group, 60% of detected aberrations in ctDNA had an allelic frequency less than 4%, which is below the threshold at which DNA sequencing at our institution can detect these same genetic alterations in tumors (26). Management of such patients can be challenging, as tissue testing is considered the gold standard.
Another source of discordance between tissue and plasma may be tumor heterogeneity. Emerging data indicate that resistance to a TKI can occur in a heterogeneous fashion (15). Biopsy of a progressing lesion, for example, may reveal a different resistance genotype than a relatively indolent lesion. Given the inherent difficulty in obtaining tissue biopsies, most trials that require biopsies for enrollment do not specify that the biopsy must be from an actively growing lesion. Thus, we do not know if the tissue biopsies obtained in the Oxnard series, for example, were from the most accessible disease site (as is usually the case) or from a progressing lesion. Another potential explanation for discordant results is differential ctDNA secretion based upon the nature of the mutation. A recent study revealed that tumor subclones harboring classical oncogenic driver mutations (e.g., EGFR) had a higher rate of ctDNA secretion than mutations in genes associated with the cell cycle (31). Further research is required to clarify the origins, significance and management of discordant results.
Serial testing
The detection of mutations by ctDNA may have utility beyond guiding selection of targeted therapies for patients. In patients with known targetable mutations on appropriate therapy, serial testing of ctDNA for the mutation can help assess treatment response. A study by Marchetti et al., for example, assessed ctDNA in patients with EGFR mutant NSCLC at baseline and then serially throughout therapy. They observed that patients who experienced a greater than 50% decrease in the allelic fraction of the EGFR variant within 14 days of treatment initiation had a better response to treatment. They also noted that absence of plasma clearance of the EGFR variant at 60 days was associated with the development of T790M mutations, and thus likely resistance to 1st and 2nd generation TKI’s (32). Subsequent studies have confirmed that clearance of EGFR variant ctDNA is associated with improved cancer outcomes (22,33).
In a study of patients with BRAF V600 mutated malignancies receiving targeted therapies, Janku et al. found that those patients who were plasma negative/tissue positive for BRAF V600 at baseline had a longer time to treatment failure (TTF) than those patients with circulating BRAF V600 on plasma testing. They also revealed that those patients who had a reduction in BRAF V600 ctDNA with treatment had a longer TTF than those whose plasma BRAF V600 levels rose or did not change (34). Serial testing may also be useful in an individual patient; Peled et al. recently described a case in which a patient’s tumor harbored multiple mutations in EGFR. The patient was treated with osimertinib for T790M positive disease, and enjoyed an excellent response aside from a progressive liver lesion. ctDNA levels of T790M fell, but the allelic fraction of a rarer EGFR mutation (G724S) increased. A biopsy of the liver lesion confirmed this tissue was T790M negative and G724S positive. Given an overall positive response to osimertinib and the documented sensitivity of G724S to afatinib, the patient was started on combination osimertinib/afatinib. The ctDNA level of G724S decreased substantially as the patient responded (35). Finally, emerging data indicate that changes in ctDNA may precede radiographic or clinical changes (17,21). Indeed, one patient described by our group experienced ctDNA progression 3 months prior to radiographic progression (26). Whether one should use progression detected by ctDNA as an indicator to alter therapies before radiographic progression remains an open question.
Another clinical space where serial testing may be useful is among patients who have completed definitive intent therapy. In a recent series by Abbosh et al., patients with lung cancer were followed with serial ctDNA analysis from diagnosis to death. They found that detection of a ctDNA heralded relapse in 13/14 patients. In addition, they found that an increase in ctDNA while on adjuvant chemotherapy was correlated with resistance and early relapse. In contrast, decreased (and undetectable) ctDNA with adjuvant chemotherapy was associated with prolonged disease-free survival (28). Another recent cohort of 41 patients with NSCLC treated with curative intent treatment confirmed the potential prognostic role of ctDNA. The detection of ctDNA had a 100% positive predictive value for disease progression, whereas the absence of ctDNA had a 93% negative predictive value for the absence of progression (36). These data imply that serial testing for ctDNA after definitive intent therapy may allow clinicians to more accurately counsel patients.
Prognostic value of ctDNA concentrations
Although the ability to detect molecular alterations in ctDNA currently has the most clinical utility, the earliest studies examined the prognostic value of the total concentration of cfDNA. In 2004 Gautschi et al. looked at plasma and serum cfDNA in NSCLC patients before the start of chemotherapy and after 1–2 cycles of chemotherapy. Increased cfDNA level after chemotherapy was associated with poor survival (37). Cargnin et al. published a meta-analysis of studies looking at cfDNA concentrations and clinical outcomes in lung cancer patients. They included 16 studies (1,723 patients) in the overall survival (OS) analysis and 5 studies (640 patients) in the PFS analysis. A higher baseline cfDNA concentration correlated with a statistically significant increase in the risk of death (HR 1.76, P<0.001). They were not, however, able to reach consensus on a cfDNA concentration cutoff point that portends a higher risk of poor outcomes given the heterogeneity of tests and reference genes that were used to detect the ctDNA (38).
In the study done by our group, a cfDNA concentration greater than 3 ng/µL was significantly associated with decreased OS; median OS for cfDNA greater than 3 ng/µL was 24 versus 46 months for cfDNA less than 3 ng/µL (P<0.01). This result remained significant when adjusted for age, performance status, EGFR mutation status, and number of metastatic sites (26).
Role of ctDNA in immunotherapy
Immunotherapy is changing the treatment landscape for NSCLC. Currently available biomarkers for the response to immunotherapy such as programmed-death ligand 1 (PD-L1) staining have significant limitations (39). Nonsynonymous tumor mutation burden (TMB) is emerging as a more reliable biomarker of response to immunotherapy in multiple tumor types, including NSCLC (40,41). Given the limitations of tissue samples in lung cancer, there is often not enough tumor to perform clinically validated tests. If mutational burden could be assessed by sampling ctDNA, this would be a great advance towards patient selection in immunotherapy. This is particularly complex though, as the gold standard for TMB analysis is whole exome sequencing. Whole exome sequencing is not feasible using current ctDNA technology. A recent study of 23 patients with advanced NSCLC revealed that in a preliminary analysis TMB in ctDNA was associated with responses to immunotherapy (42).
Use of ctDNA in lung cancer screening
The National Lung Screening Trial found that annual low dose computed tomographic (CT) scans of the chest decrease mortality among patients at high risk for the development of lung cancer (43). False positive results on CT scans, however, represent a significant limitation of screening. There has been interest in applying cfDNA technology in order to improve the accuracy of lung cancer screening, but this has not been borne out in subsequent clinical trials. In 2009 Sozzi et al. enrolled 1,025 heavy smokers to receive annual low-dose spiral CT along with serial plasma cfDNA quantification by PCR over 5 years. Assessment of plasma DNA did not improve the accuracy of lung cancer screening—those patients who went on to develop cancer did not have higher cfDNA levels than those who did not develop cancer during the study (44). One potential limitation to using cfDNA levels to help detect early lung cancers is that cfDNA levels can be increased in patients with conditions other than lung cancer, such as liver disease, diabetes, cardiovascular disease, non-neoplastic lung diseases and infections (45,46).
Conclusions
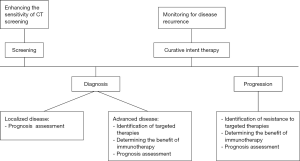
Treatment paradigms in advanced NSCLC have changed considerably in recent years; targeted and immune directed therapies provide new hope in an otherwise lethal disease. As we find more ways to target the mutations driving tumor growth, we will need better access to tumor genetic materials that can help guide clinical decisions. ctDNA has emerged as a potential solution to this dilemma. Given the superior outcomes of patients on appropriate targeted therapy in advanced NSCLC this advance in personalized treatment holds great promise. To validate the analysis of ctDNA for more widespread use, we will need continued refinement and standardization of the technology as well as rigorous prospective clinical trials. In Figure 1, we describe how cfDNA may be applied in the management of NSCLC.
Acknowledgements
None.
Footnote
Conflicts of Interest: C Aggarwal: Consultant Services: Roche, BMS, Eli Lilly; Research/Grant support: Takeda, Macrogenics, Roche. E Carpenter: Commercial research support from Janssen/Johnson & Johnson. RB Cohen: Consultant Services: Takeda, Zymeworks, BMS. CJ Langer: Consultant services: Bristol-Myers Squibb Company; ImClone Systems Incorporated; Pfizer Inc.; Eli Lilly and Company; AstraZeneca Pharmaceuticals LP; Novartis Pharmaceuticals Corporation; Genentech, Inc.; Bayer HealthCare Pharmaceuticals/Onyx Pharmaceuticals, Inc.; Celgene; Abbott Laboratories; Biodesix; Clariant; CarisDx; ARIAD Pharmaceuticals, Inc.; Boehringer Ingelheim Pharmaceuticals, Inc.; Synta Pharmaceuticals Corp; Clovis Research/grant support: Pfizer Inc.; Eli Lilly and Company; GlaxoSmithKline; Clovis; Merck; Nektar; Advantagene; Inovio; Ariad; Celgene; DSMC member: Amgen, Synta, Peregrine, SWOG, Incyte, Lilly CME: PIK; PER; NOCR; CCO; RTP, MLG. J Bauml: Consultative Services: Clovis, BMS, Astra Zeneca, Celgene, Merck, Genentech, Guardant Health, Boehringer Ingleheim; Research/Grant Support: Merck, Incyte, Carevive Systems, Novartis, Bayer. M Marmarelis, JC Thompson and TL Evans have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, et al. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev 2015;41:361-75. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The lancet oncology 2012;13:239-46. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced alk-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw 2014;12:1738-61. [Crossref] [PubMed]
- Spira A, Halmos B, Powell CA. Update in Lung Cancer 2014. Am J Respir Crit Care Med 2015;192:283-94. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFR T790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Chandrananda D, Thorne NP, Bahlo M. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med Genomics 2015;8:29. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. [Crossref] [PubMed]
- Dressman D, Yan H, Traverso G, et al. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A 2003;100:8817-22. [Crossref] [PubMed]
- Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res 2015;21:3631-9. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 2015;10:e0140712. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772-82. [Crossref] [PubMed]
- Caruso R, Parisi A, Bonanno A, et al. Histologic coagulative tumour necrosis as a prognostic indicator of aggressiveness in renal, lung, thyroid and colorectal carcinomas: A brief review. Oncol Lett 2012;3:16-8. [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Wakelee HA, Gadgeel SM, Goldman JW, et al. Epidermal growth factor receptor (EGFR) genotyping of matched urine, plasma and tumor tissue from non-small cell lung cancer (NSCLC) patients (pts) treated with rociletinib. J Clin Oncol 2016;34:abstr 9001.
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016.JCO667162. [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Janku F, Huang HJ, Claes B, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther 2016;15:1397-404. [Crossref] [PubMed]
- Peled N, Roisman LC, Miron B, et al. Subclonal Therapy by Two EGFR TKIs Guided by Sequential Plasma Cell-free DNA in EGFR-Mutated Lung Cancer. J Thorac Oncol 2017;12:e81-e4. [Crossref] [PubMed]
- Chaudhuri A, Chabon JJ, Lovejoy AF, et al. Analysis of circulating tumor DNA in localized lung cancer for detection of molecular residual disease and personalization of adjuvant strategies. J Clin Oncol 2017;35: abstr 8519.
- Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic Acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004;22:4157-64. [Crossref] [PubMed]
- Cargnin S, Canonico PL, Genazzani AA, et al. Quantitative Analysis of Circulating Cell-Free DNA for Correlation with Lung Cancer Survival: A Systematic Review and Meta-Analysis. J Thorac Oncol 2017;12:43-53. [Crossref] [PubMed]
- McLaughlin J, Han G, Schalper KA, et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:46-54. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Cai W, Zhou C, Su C, et al. MA15. 03 The Predictive Value of Mutation/Neoantigen Burden from ctDNA on the Efficacy of PD-1 Blockade in Advanced NSCLC. J Thorac Oncol 2017;12:S429-S30. [Crossref]
- National Lung Screening Trial Research T, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med 2009;179:69-74. [Crossref] [PubMed]
- Casoni GL, Ulivi P, Mercatali L, et al. Increased levels of free circulating DNA in patients with idiopathic pulmonary fibrosis. Int J Biol Markers 2010;25:229-35. [PubMed]
- Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann Transl Med 2014;2:107. [PubMed]


