Emerging uses of biomarkers in lung cancer management: molecular mechanisms of resistance
Introduction
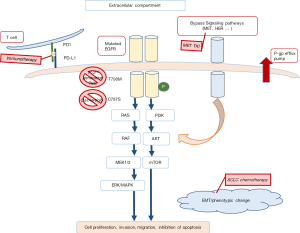
Research over the last decade has transformed the management of patients with advanced non-small cell lung cancer (NSCLC) with the recognition of molecularly defined subsets of tumors with unique sensitivities to targeted therapeutics, such as patients with EGFR/ALK/ROS-mutated lung adenocarcinoma. Upfront molecular therapy is now the standard of care for optimization of treatment, and patient outcomes have greatly improved. In addition, an expanding group of other molecular alterations are continuing to be recognized and biomarker-driven immunotherapeutic strategies have yielded dramatic advances in patient management. However, acquired resistance to both targeted and immunotherapeutic agents have become a pivotal issue limiting the long-term benefit of such therapies. In the current review, we will highlight the most significant advances in our understanding of mechanisms of primary and acquired drug resistance, strategies to detect secondary molecular events, and drug development strategies yielding new generations of inhibitors with increasing success to overcome acquired resistance (Figure 1).
Epidermal growth factor receptor (EGFR)
The EGFR gene, located on chromosome 7p12-13, encodes for a HER family receptor tyrosine kinase (RTK) that upon activation, will “switch on” several downstream signaling pathways important in cell survival and proliferation (1). Activating mutations in the tyrosine kinase domain of the EGFR gene are seen in 10–15% of non-squamous NSCLC and are responsible for tumor growth, proliferation, invasion and metastases by promoting pro-proliferative and anti-apoptotic effects. These mutations occur more commonly in tumors in women, non-smokers and patients of Asian ethnicity and can be as frequent as 50–60% in lung adenocarcinomas in Asian women (2,3).
Two so-called “classic” mutations account for >90% of the known activating EGFR mutations: L858R point mutation on exon 21 and in-frame deletions around the conserved LREA motif of exon 19 (2,3). The presence of such activating EGFR mutations serves as a biomarker as well as a target for therapy with EGFR-tyrosine kinase inhibitors (TKI). Several clinical studies revealed that response rates (RRs) to first generation EGFR-TKIs, such as gefitinib and erlotinib, in patients harboring an EGFR-activating mutation are 50–80% and can lead to durable responses with progression free survival in the 8–12 months range (4-8). In 2012, a meta-analysis of six randomized controlled trials confirmed significant improvement in overall response rate (ORR) and doubling of progression free survival (PFS) in patients with advanced EGFR mutated NSCLC receiving first-line EGFR-TKI, as compared to conventional chemotherapy (9). Therefore, EGFR-TKIs are the current standard treatment for these patients and frontline molecular testing is now part of routine management (4,9).
However, resistance to EGFR-TKIs poses a major clinical problem, and can be categorized as primary or acquired. Primary resistance refers to de novo lack of response to the targeted therapy, while acquired resistance is defined as progression of disease after an initial period of clinical response (10,11).
Primary resistance
Several mechanisms of primary resistance to EGFR-TKIs have been described.
First generation EGFR-TKIs are not effective in patients with the gatekeeper EGFR T790M mutation which can be present as a germline mutation in rare familial clusters. This mutation will be discussed in detail later in this review (10).
Another example of primary resistance to EGFR TKIs is the EGF exon 20 insertion mutation. The RR of NSCLCs with EGFR exon 20 insertion mutation to EGFR-TKIs is below 5% (12). The mechanism for this primary resistance has been described by Yasuda et al. (12), 80–90% of activating EGFR exon 20 mutations cause insertion of one to four amino-acids beyond the C-helix of the tyrosine kinase domain, forming a wedge at the end of the C helix that promotes the active kinase and leaves the adenosine triphosphate binding pocket unaltered. Interestingly, an EGFR exon 20 insertion mutation sensitive to the EGFR-TKIs was also identified (EGFR A763_Y764insFQEA). Structurally, this mutation is very different from other exon 20 insertions and more closely resembles the L858R and exon 19 deletion mutations. At present, the standard of care for TKI-resistant exon 20 mutated EGFR tumors is conventional chemotherapy (13,14). Efforts are being made to develop EGFR targeting agents with activity against this important class of mutation (e.g., AP32788 NCT02716116).
Another mechanism of primary resistance is conferred by polymorphisms in the Bim gene. Bim is a potent pro-apoptotic protein and its expression is suppressed in EGFR mutated lung cancers. TKI therapy can upregulate Bim expression and allow for cell death and tumor regression. In the case of certain inherited Bim gene polymorphisms, the pro-apoptotic domain is not present and TKI therapy is less successful with shorter duration of response than in other genotypes (15).
Finally, as expected, EGFR-TKI therapy is not effective against tumors with concurrent activating mutations in genes downstream of EGFR such as K-ras and BRAF or against tumors harboring other oncogenic gene alterations (10). Canale et al. (16) observed that TP53 tumor suppressor gene mutations, especially exon 8 mutations, reduce the RR to TKIs. Inferior outcomes in the presence of p53 mutations had been similarly noted by the Lung Cancer Mutation Consortium (17). TP53 mutations occur in about 30%–40% of NSCLCs and while TP53 mutations might not be actionable, this may be important in order to risk stratify patients.
Acquired resistance
Acquired resistance to EGFR-TKIs typically develops within 6–18 months of starting therapy (11). Biopsy and sequencing of tumors in patients with disease progression on an EGFR TKI as well as in vitro and mouse studies have led to the discovery of potential mechanisms of acquired resistance as well as novel and effective ways of overcoming certain resistance mechanisms (18).
T790M mutation
The most common mechanism of acquired resistance, accounting for up to 60% of cases, is a secondary mutation of EGFR gene, leading to the substitution of methionine for threonine at position 790 (T790M) (10). Methionine’s large side chain causes steric hindrance and reduces the ability of 1st generation EGFR-TKIs such as erlotinib and gefitinib to bind to the ATP-kinase pocket. In addition, this mutation changes the dynamics at the binding site such that ATP, rather than the ATP-competitive EGFR-TKIs is the favored substrate. This leads to a 1,000-fold increased resistance against the EGFR-TKIs (10,18).
Several studies have been done to determine if tumor cells containing the T790M mutation are present prior to EGFR-TKI treatment initiation or if they develop from cancer stem cells during treatment. These studies have found very few TKI-resistant cells prior to treatment thereby suggesting that the T790M mutation more commonly arises due to selective pressure during EGFR-TKI therapy (10,19). Three other point mutations have been implicated in EGFR-TKI resistance (D761Y, L747S, T854A), however they have been reported only occasionally and their mechanism of resistance is less understood (10).
Activation of bypass signaling pathways
A second mechanism of acquired resistance is through the activation of bypass signaling pathways. For example, amplification of the MET gene has been seen in 5–22% of NSCLC patients who develop acquired resistance to EGFR-TKIs. These amplifications are seen in very low rates in tumors that have not yet undergone treatment with an EGFR-TKI. Thus, it has been postulated that under the selective pressure of EGFR-TKI therapy, cells with dual EGFR and MET activation can undergo clonal selection. Ultimately, these EGFR + MET mutated cells form the bulk of the tumor leading to clinical resistance against EGFR-TKIs. In vitro, MET amplification has been reported in gefitinib-resistant cell lines and dual EGFR and MET inhibition has been successful in inducing apoptosis in this case (20,21).
MET activation by its ligand HGF may also be another mechanism contributing to acquired resistance. In a few studies, it has been noted that HGF is expressed in high levels in 29% of patients with primary resistance and 61% of patients with acquired resistance (22). If future studies validate this mechanism in vivo, targeted therapy against HGF can be a potential strategy to treat this subset of patients (10).
HER2 amplification is another alternative signaling pathway that plays a role in acquired resistance to EGFR-TKIs and has been noted in up to 13% of patients (10). In a phase Ib study of patients with NSCLC resistant to erlotinib/gefitinib, dual inhibition of EGFR and HER2 with afatinib and cetuximab demonstrated promising activity leading to median PFS of 4.7 months (23). Another bypass signaling mechanism that has been recently implicated in afatinib-resistant PC9 cells is the IGF1R pathway—increased expression of insulin-like growth factor binding protein 3 (IGFBP-3) enhances IGF1R activity leading to increased AKT phosphorylation and subsequent cell cycle progression (24).
Phenotypic changes
Yet another mechanism of acquired resistance is through phenotypic alterations. Epithelial mesenchymal transition (EMT) is defined as the loss of epithelial markers and a subsequent gain of mesenchymal features (10). NSCLC cells that undergo EMT lose sensitivity to EGFR-TKIs (25). Furthermore, when the epithelial phenotype is restored in these EGFR-mutated NSCLC cell lines, they become sensitive again to EGFR-TKIs (26). The transition between epithelial and mesenchymal phenotypes is likely influenced by the AXL RTK. A high level of expression of AXL RTK has been implicated in acquired resistance to erlotinib, and over-expression of this same RTK has been noted in NSCLC cells with the mesenchymal subtype (10,27). Taken together, it seems as though AXL plays a significant role in the acquired resistance to EGFR-TKIs and may one day be an effective pharmaceutical target. In fact, a recent report of tumor genomic profiling cites the case of a patient with lung adenocarcinoma and pleural carcinomatosis whose genome analysis showed focal gain of chromosome 19q12-13.11, including AXL. He was enrolled in a Phase I trial of MGCD265, a TKI targeting MET and AXL with a dramatic response and near resolution of lung infiltrates on imaging in just 2 months (28).
A different kind of phenotypic alteration happens when the NSCLC cells undergo histological transformation to small cell lung cancer (SCLC). This has been reported in 3–14% of patients with acquired resistance to EGFR-TKI who underwent re-biopsy. In a recent study Lee et al. (29) investigated 21 patients with advanced EGFR-mutated NSCLC that transformed into SCLC. Whole genome sequencing was performed at different time points during disease evolution. It was concluded that TKI-resistant EGFR-mutated NSCLC and SCLCs share a common clonal origin and the clonal divergence occurs before the EGFR TKI therapy. Complete inactivation of RB1 and TP53 were observed from the early NSCLC stages in these tumors. There have been multiple cases that reported good response to SCLC chemotherapy regimens once the NSCLC undergoes this histological transformation underlining the importance of testing for such alterations (10,30).
Pharmacodynamic limitations
Another limitation to the use of first and second generation TKIs is the inability of these drugs to fully penetrate the blood brain barrier (BBB) and enter the central nervous system (CNS). Patients can often develop CNS metastases during treatment, even when their extracranial tumors are still under control (31). CNS concentrations of all currently available agents are lower than plasma [gefitinib has a CSF concentration approximately 1% of serum and erlotinib has a CSF concentration 5% of serum (32,33)]. Many EGFR TKIs that have been designed for improved CNS penetration are currently under investigation. One such drug, AZD3759 has shown promise with CNS penetration adequate enough to promote tumor shrinkage in patients with brain and leptomeningeal metastases in addition to a tolerable side-effect profile (34).
Treatment approaches to overcome resistance
Third generation EGFR TKIs
Knowledge of the biological mechanisms behind acquired resistance to EGFR-TKIs has led to the development of 3rd generation TKIs that specifically target the EGFR T790M mutant cells. The side-effect profile of these new TKIs is more favorable given that they spare WT EGFR (10,11).
Of the 3rd generation TKIs, osimertinib has progressed the furthest in clinical development. Osimertinib, an oral irreversible agent with CNS penetration, had initially been approved by the FDA under the Breakthrough Therapy Designation Program based on outstanding results from the AURA-1 and AURA-2 studies suggestive of great efficacy with RRs in the 50–60% range in EGFR T790M-mutated cases as well as favorable toxicity profiles. Recently, Mok et al. published the results of a confirmatory, randomized, open-label, international, phase 3 (AURA 3) trial to show the superiority of osimertinib over platinum plus pemetrexed chemotherapy in patients with confirmed T790M-positive advanced NSCLC after first-line EGFR-TKI therapy. Median PFS was significantly longer with osimertinib as compared to chemotherapy (10.1 vs. 4.4 months) and objective RR was significantly better with osimertinib (71% vs. 31%). Even among the 144 patients in the trial who had CNS disease, osimertinib performed better, PFS was 8.5 months with osimertinib vs. 4.2 months with chemotherapy. Adverse events of grade 3 or higher were more common in the chemotherapy group as compared to osimertinib (47% vs. 23%) (35). These results led to full FDA approval of osimertinib in 2017.
Unfortunately, tumor samples from patients treated with osimertinib have already revealed that emerging resistance will remain a key shortcoming of this class of drugs. The EGFR C797S mutation appears to be the most dominant resistance mechanism, with frequency as high as 20–40% (36). This mutation is very logical from a biochemical perspective as osimertinib and other irreversible EGFR inhibitors covalently bind to C797 of EGFR gene and this mutation interferes with such binding, thus drastically diminishing activity. Interestingly, the allelic context of C797S seems to have treatment implications—when the T790M and C797S mutations occur in trans (on separate alleles), a combination of first and third generation TKIs can restore EGFR inhibition. But, if the mutations are in cis (on same allele), the cells will not be sensitive to any available EGFR TKI (37). Understanding of such novel resistance mechanisms is already leading to innovative approaches of preventing/overcoming such resistance.
For example, a new compound called EAI045, which acts as an allosteric inhibitor of the TK domain of EGFR, could be a promising agent in overcoming resistance to 3rd generation TKIs. In mouse models, EAI045 in combination with cetuximab, an antibody that stops EGFR dimerization, has been shown to be effective against the L8585R/T790M and L858R/T790M/C797S EGFR mutants (38). Another drug regimen that is a powerful candidate to overcome triple-mutant EGFR is the combination of cetuximab with brigatinib, a potent and selective ALK/EGFR T790M inhibitor. This regimen showed excellent efficacy in del19/T790M and del19/T790M/C797S cell lines as well as in del19/T790M/C797S xenograft mouse models (39).
MET inhibition for bypass signaling
Dual inhibition of EGFR and MET has been studied in two Phase III trials: the MET-Lung trial that investigated the combination of erlotinib ± onartuzumab, a monoclonal antibody against MET, as well as the MARQUEE trial that compared erlotinib ± tivantinib, a MET TKI. However, neither of these studies met their primary endpoint of improving OS, maybe due to the fact that the studies did not focus on the EGFR-mutated population (10,40).
Since successful treatment with erlotinib and crizotinib (a potent MET and ALK inhibitor) has been described in case reports (41,42), a phase I trial was designed to study this combination. However, in the 26 patients in the trial, the MTD for the two drugs was lower than the approved dose of the individual drugs, so phase II trials have not been initiated yet (43). Further trials have also been halted for the combination regimen of erlotinib ± the anti-MET antibody LY2875358 because of initial disappointing results (10).
There are more promising biomarker-driven studies that are currently being pursued. Preliminary studies of another MET inhibitor, capmatinib revealed that patients with cMET gene copy number ≥5 had an ORR of 40% (44). At present, a three-arm study has been designed to study capmatinib by itself and in combination with erlotinib as compared to standard chemotherapy in EGFR-mutated NSCLC patients specifically with c-MET copy number >6 (NCT02468661).
Another study will compare the combination of gefitinib and a MET inhibitor named tepotinib versus chemotherapy as second line therapy in patients with MET + EGFR mutant tumors with acquired resistance to gefitinib (NCT 01982955). The phase IB data from this study revealed 4/18 partial responses and a good side-effect profile, and phase II is currently in process (45).
Volitinib is another MET inhibitor that is presently being studied. One trial explores the safety of the volitinib + gefitinib combination in EGFR-mutated NSCLC (NCT02374645), while the TATTON trial is investigating volitinib in combination with different doses of osimertinib in EGFR-mutant NSCLC that has progressed on prior EGFR-TKI therapy (NCT02143466). Phase Ib data from the TATTON study was promising, demonstrating partial responses in 6/11 patients (46).
In all these studies that involve MET-targeting drugs, it is imperative to have assays that reliably detect MET pathway activation and alterations and serve as biomarkers that predict response to MET inhibitors. Currently, there are a number of methods that can be used to determine MET expression, however not all of them are equal. For example, immunohistochemistry (IHC) is an easy assay but studies have revealed that it is a poor marker of predicting sensitivity to MET inhibitors (47). Fluorescent in situ hybridization (FISH) and rtPCR are reliable methods for detecting copy number alterations in the MET gene, although the latter encounters problems in distinguishing polysomy from true copy number changes (48). One drawback with FISH was the question of what cutoff should be used to predict sensitivity to MET inhibitors. A recent study by Noonan et al sheds light on the matter as it showed that a FISH MET/CEP7 ratio of ≥5 was associated with the most sensitivity to MET inhibition (49). While circulating-tumor DNA (ctDNA) testing might be a good option for capturing MET point mutations, however it is less sensitive to detect copy number changes and gene rearrangements. Perhaps the most effective method of detecting MET gene alterations and predicting response to MET inhibitors is next-generation sequencing (NGS). This is because NGS has the ability to reveal the multitude of genomic changes impacting the MET gene.
A range of studies focusing on novel combination treatments, in an effort to develop appropriate synergistic treatment strategies, for example combinations of immunotherapeutic, anti-angiogenic and Met/AXL/MEK targeting agents, are ongoing (Table 1).
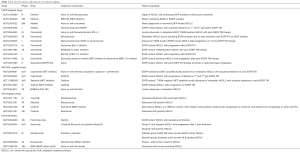
Full table
Other oncogenes
Anaplastic lymphoma kinase (ALK)
ALK gene chromosomal rearrangements define another distinct molecular subtype of NSCLC. Therapeutic options for the treatment of advanced ALK-positive NSCLC have expanded rapidly through the development of increasingly potent and selective ALK inhibitors, however the problem of developing resistance to these therapies have emerged as a key clinical issue. Efforts to understand the mechanisms of resistance to ALK inhibitors have led to recognition of many different mechanisms of resistance.
Crizotinib, the first generation ALK inhibitor, significantly improves the objective response rates (ORRs) and progression-free survival (PFS) compared to cytotoxic chemotherapy in advanced ALK-positive lung cancer and has become the standard front-line therapy for ALK-positive patients. However, most patients who initially respond to crizotinib develop progressive disease within 1 year (50-52). Initial studies examining biopsies obtained at the time of resistance revealed multiple potential resistance mechanisms. Mechanisms of resistance to ALK inhibitors are divided into two categories, on target or ALK dependent alterations and off target or ALK independent alterations. On target alterations include mutations in the ALK tyrosine kinase domain and amplification of ALK (53,54).
Mutations in the ALK tyrosine kinase domain can cause resistance to ALK inhibitors by directly blocking the TKI binding to the target kinase, altering the kinase’s conformation, or altering the ATP-binding affinity of the kinase. Many mutations in the ALK tyrosine kinase domain have been identified. For example, the L1196M gatekeeper mutation alters the gatekeeper residue at the bottom of the ATP-binding pocket and impairs TKI binding; and solvent-front mutations impair drug binding likely through steric hindrance. Interestingly each ALK TKI appears to be associated with a distinctive pattern of ALK resistance mutations (53,54). Amplification of ALK occurs less frequently and causes resistance to crizotinib. It has been reported in 6.7% to 18.2% of cases of resistance to crizotinib in different series (51,55). ALK amplification has not yet been detected as a resistance mechanism after second generation ALK TKIs (53).
Similar to the case of acquired EGFR TKI resistance, off target alterations that cause resistance to ALK inhibitors include activation of bypass signaling pathways, lineage changes and drug efflux pump (53,54). Numerous examples of bypass signaling activation have been discovered. For example, gene expression profiling of crizotinib-resistant versus crizotinib-naive NSCLC tumor samples identified EGFR and HER2 signatures as two of the most enriched gene expression signatures in resistant tumors (51,56,57). In one study, increased EGFR activation was identified in 4 of 9 cases (44.4%) when biopsy samples prior to crizotinib initiation and after development of resistance were compared (51). This was associated with higher EGFR mRNA expression and persistent activation of downstream ERK and AKT signaling. EGFR activation may result from receptor and/or ligand upregulation rather than EGFR mutations or amplification (51,58). MET amplification is another example of bypass signaling. Crizotinib is a potent ALK and MET inhibitor however most other ALK inhibitors do not have anti-MET activity. MET amplification has recently been reported as a bypass mechanism in patients who have failed second-generation inhibitors like alectinib (59-61). Direct activation of downstream signaling pathways can also enable acquired resistance to ALK inhibitors. MEK activating mutations, PIK3CA mutations, KIT amplification, IGF1R activation, SRC activation and TP53 mutations have all been identified, among others (62-64). Of note, ALK/MEK dual blockade may be effective not only in overcoming but also in delaying ALK TKI resistance and are being tested in early phase clinical trials (63,65). Similar to EGFT TKI resistance, phenotypic changes such as EMT and SCLC transformation might cause ALK TKI resistance (53,66).
Lastly, P-glycoprotein (P-gp) is an ATP-dependent efflux pump encoded by the multidrug resistance 1 (MDR1) gene (67) and might mediate cases of pharmacodynamics resistance. P-gp is a substrate for crizotinib and ceritinib and its overexpression can lead to acquired ceritinib and crizotinib resistance by exporting the drug outside of the cancer cells. P-gp has also been implicated in decreased penetration of the blood–brain barrier by P-gp. Alectinib is not a P-gp substrate (54,68).
Second-generation ALK inhibitors, ceritinib and alectinib, were rapidly developed and showed excellent activity in the second line setting, leading to FDA approval of both compounds (69,70). ORRs to second-generation ALK inhibitors are reported to be 48% to 71% following progression on crizotinib (71,72). A third agent, brigatinib was recently granted accelerated approval for the treatment of patients with ALK positive NSCLC who have progressed on or are intolerant to crizotinib (73,74). Intriguingly, the activity of these agents seems to be similar regardless of the presence or the absence of secondary ALK mutations and therefore repeat mutation testing at the time of crizotinib resistance has less clear value as compared to for example EGFR T790M mutation testing. It appears that part of the reason for this is the relatively weak ALK inhibitory potency of crizotinib as compared to these novel agents.
Recent data is more supportive of the need for molecular testing at the time of acquired resistance with the availability of several more potent agents as the type of emerging mutation might be a key determinant of efficacy of other alternate ALK inhibitors.
In a recent article, Gainor et al. (53) reported mechanisms of resistance in 83 ALK-positive patients who underwent repeat biopsies following disease progression on first or second generation ALK inhibitors. One hundred and three repeat biopsy samples from 83 patients were evaluated using genetic and cell based assays. ALK kinase domain mutations were found in 20% (11/55) of crizotinib resistant samples. The two most common mutations were the L1196M (gatekeeper mutation) in 7% and the G1269A in 4% of patients. ALK kinase domain mutations were detected in 54% (13/24) of ceritinib resistant specimens, 53% (9/17) of alectinib resistant samples, and 71% (5/7) of brigatinib resistant samples. Distinct patterns of mutations were found for each of the three second-generation ALK inhibitors. The G1202R solvent front mutation, which produces resistance to 1st and 2nd generation ALK inhibitors, was found in 21% ceritinib, 29% alectinib, and 43% of brigatinib resistant samples. This has clinical implications as the novel ALK inhibitor, lorlatinib can overcome this mutation. Compound mutations were identified in 12.5% (6/48) of patients resistant to second-generation ALK TKIs. In vitro, lorlatinib was the only agent to retain potency against the compound mutations evaluated. EMT was observed in 42% (5/12) of cases. The complex evolution of resistance mutations over multiple lines of ALK TKI therapy and the potential for optimizing drug selection based on the understanding of secondary ALK mutations reinforces the need for repeat biopsies to tailor therapeutic selection and ctDNA is also being developed as a robust platform for molecular monitoring (75).
ROS/MET/BRAF
ROS
Patients with ROS proto-oncogene 1 RTK (ROS1) rearranged NSCLC in general respond to treatment with the ROS1 inhibitor-crizotinib very well with quite durable remissions being the norm, however these tumors eventually become resistant as well (76,77). There is limited data available on the mechanisms of acquired resistance to crizotinib in ROS1 positive NSCLC, however several secondary ROS1 mutations that can cause resistance have been identified, for example the G2026M mutation (gatekeeper) or the L2155S solvent-front (D2033N) mutation which can still respond to the multi-targeted TKI, cabozantinib which has ROS inhibitory activity as well (78-80). Other mechanisms of resistance might include gain of function mutations of KIT (81), activation of the RAS pathway due to either KRAS/NRAS mutations or to KRAS amplification (82), rat sarcoma viral oncogene homolog, EGFR activation (83), and epithelial-to-mesenchymal transition (78,84).
MET
NSCLC harboring MET mutations such as MET exon 14 skipping mutations or focal amplification can respond to treatment with MET TKIs, such as crizotinib and cabozantinib. Acquired resistance to MET TKIs have been reported, however the molecular mechanisms are not yet well defined (85). In a recent study, Li et al. (86) reported two acquired MET mutations, Y1248H and D1246N, which can cause resistance against Type I MET-TKIs. It was also noted that EGFR amplification may act as an alternative MET-TKIs bypass resistance mechanism (86). Amplification of MET, amplification of K-RAS and activation of B-RAF or EGFR pathways have also been proposed as mechanisms of acquired resistance to MET TKIs (87-89). Knowledge of specific molecular mechanisms might yield treatment benefits as some secondary MET mutations might respond to for example type II MET TKIs.
BRAF
In V600 BRAF mutated NSCLC, duration of response to BRAF TKIs are in general relatively short due to the development of resistance. Resistance patters are diverse and can occur due to a secondary MAPK alteration or due to activation of bypass tracks (90,91). Non-V600E BRAF mutations demonstrate limited sensitivity to treatment with BRAF TKIs and studies are ongoing to assess the utility of MEK inhibition in such cases (92).
Mechanisms of resistance to BRAF are much better studied in patients with malignant melanoma.
For instance, Johnson et al. (93) studied 132 tumor specimens from 100 patients with malignant melanoma who had developed resistance to BRAF inhibition, the mechanisms of resistance included NRAS or KRAS mutations in 20%, BRAF splice variants in 16%, BRAF amplifications in 13%, MEK1/2 mutations in 7%, and non-mitogen-activated protein kinase pathway alterations in 11%. While these resistance mechanisms are likely shared with V600-mutated lung cancers, this will still certainly need to be demonstrated.
ctDNA as a novel testing platform in the monitoring of acquired resistance
CtDNA analysis has emerged as a powerful, and in certain settings validated, tool that allows clinicians to non-invasively detect actionable mutations (94,95). EGFR T790M testing provided the first validated usage for ctDNA testing and while tissue biopsy remains the gold standard for EGFR T790M genotyping, ctDNA is emerging as a valuable alternative and/or complementary tool. Zheng et al. reported 81.25% sensitivity and 100% specificity of plasma T790M testing by dPCR assays in a study of 117 patients with TKI resistance (96). Oxnard et al. demonstrated that the sensitivity of ctDNA detection of T790M was 70% and the objective RR and median PFS with osimertinib were similar in patients with T790M-positive plasma or T70M-positive tumor results (97). It appears that the positive predictive value of detecting EGFR mutations in ctDNA is high enough to justify initiation of 3rd generation TKI therapy, but the negative predictive value is less robust. Currently, studies are in progress to test the efficacy of ctDNA in detecting ALK resistance mutation (98).
Acquired resistance to immunotherapeutic agents
Immune checkpoint inhibitors are now widely used as single agents for treatment of patients with advanced NSCLC in front-line or later lines of therapy. PD-1/PD-L1 blockade is a revolutionary treatment option for advanced NSCLC, however there is a high rate of primary resistance, and furthermore most patients develop acquired resistance to PD-1/PD-L1 blockade (99).
Mechanisms of primary resistance to immunotherapeutic agents include lack of PD-L1 expression on tumor cells, insufficient tumor-infiltrating lymphocytes or severely exhausted CD8+ T cells (100). Another primary resistance mechanism might be the scarcity of neo-antigens in tumors with a low mutation burden, such as EGFR/ALK/ROS-mutated lung cancers where the average tumor mutation burden is a magnitude or so lower than in smoking-associated lung cancers and indeed low RRs of 4% had been reported in this subset of patients (53).
Acquired resistance, defined by initial response to PD-1 blockade followed by progression of disease, is observed commonly. Few mechanisms for acquired resistance have been clearly identified as of yet but better understanding of determinants of efficacy and resistance will be pivotal to allow optimal utilization of these novel agents. Candidate resistance mechanisms might include upregulation of alternate immune checkpoints (101), somatic mutations in HLA or JAK1/JAK2 genes (102,103) and genomic changes resulting in loss of mutation-associated neoantigens in resistant clones (104).
Koyama et al. (101) analyzed the tumor immune microenvironment in two fully immunocompetent mouse models of lung adenocarcinoma. In tumors with acquired resistance to PDL-1 blockade, upregulation of alternative immune checkpoints, notably T-cell immunoglobulin mucin-3 (TIM-3) in PD-1 antibody bound T cells was identified.
Zaretsky et al. (103) demonstrated that acquired resistance to immunotherapeutic agents is possible through genetic aberrations translating into immune escape mechanisms in tumor cells which are then clonally expanded, leading to disease relapse in initially responding patients. Whole exome sequencing of 4 paired melanoma specimens at baseline and at relapse revealed loss of function mutations in the genes encoding interferon receptor associated Janus kinase 1 (JAK1) or Janus kinase 2 (JAK2), concurrent with deletion of the wild-type allele in 2 of 4 patients. JAK1 and JAK2 mutations resulted in a lack of response to interferon gamma, including insensitivity to its anti-proliferative effects on cancer cells. A truncating mutation in the gene encoding the antigen-presenting protein beta-2-microglobulin (B2M) was identified in a third patient. The B2M truncating mutation led to loss of surface expression of major histocompatibility complex class I, which is a previously identified mechanism for cytotoxic T-cell escape. Similar results have yet to be reported in lung tumors.
Anagnostou et al. (104) performed whole exome sequencing in four patients with NSCLC who developed acquired resistance to PD-1 blockade and compared the results in pre-treatment and post-treatment specimens and identified genomic changes resulting in loss of 7 to 18 mutation associated neoantigens in resistant clones. Peptides generated from the eliminated neoantigens elicited clonal T-cell expansion in autologous T-cell cultures, suggesting that they generated functional immune responses.
More studies are needed and efforts need to be expanded to better understand mechanisms of resistant and to be able to develop more effective drugs or combination strategies to enhance efficacy of immunotherapies.
Summary
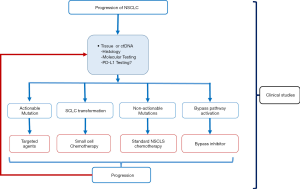
The introduction of molecular targeted and immunotherapeutic agents have completely transformed the landscape of NSCLC management, dramatically improving patient outcomes. Molecular and immune biomarker testing is paramount in optimal treatment selection and acquired resistance is a pivotal clinical issue with an increasing understanding of a range of mechanisms (Figure 2).
Molecular monitoring utilizing tissue biopsy or plasma is now a routine element of patient management with the availability of second and third-generation agents. Continued efforts towards novel combination studies to overcome and possibly prevent the emergence of acquired resistance remain the most critical experimental need.
Acknowledgements
None.
Footnote
Conflicts of Interest: B Halmos has received consulting fees from Astra-Zeneca, Boehringer-Ingelheim, Foundation One, Genoptix, Eli-Lilly, Roche, Pfizer, Takeda, Novartis and research funding from Mirati, Astra-Zeneca, Merck, Bristol-Myers-Squibb, Boehringer-Ingelheim, Takeda, Eli-Lilly, Novartis. The other authors have no conflicts of interest to declare.
References
- Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 2010;277:301-8. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Gao G, Ren S, Li A, et al. Epidermal growth factor receptor-tyrosine kinase inhibitor therapy is effective as first-line treatment of advanced non-small-cell lung cancer with mutated EGFR: A meta-analysis from six phase III randomized controlled trials. Int J Cancer 2012;131:E822-9. [Crossref] [PubMed]
- Saad S, Huang K, Halmos B. Overcoming resistance to EGF receptor tyrosine kinase inhibitors in EGFR-mutated NSCLC. Lung Cancer Management 2014;3:459-76. [Crossref]
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-59. [Crossref] [PubMed]
- Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. [Crossref] [PubMed]
- Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177. [Crossref] [PubMed]
- Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179-84. [Crossref] [PubMed]
- Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012;18:521-8. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Aisner D, Sholl LM, Berry LD, et al. Effect of expanded genomic testing in lung adenocarcinoma (LUCA) on survival benefit: The Lung Cancer Mutation Consortium II (LCMC II) experience. J Clin Oncol 2016;34:abstr 11510.
- Sharma J, Shum E, Chau V, et al. The Evolving Role of Biomarkers in Personalized Lung Cancer Therapy. Respiration 2017;93:1-14. [Crossref] [PubMed]
- Yu HA, Arcila ME, Hellmann MD, et al. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol 2014;25:423-8. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [Crossref] [PubMed]
- Yano S, Yamada T, Takeuchi S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol 2011;6:2011-7. [Crossref] [PubMed]
- Janjigian YY, Smit EF, Groen HJ, et al. Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov 2014;4:1036-45. [Crossref] [PubMed]
- Yamaoka T, Ohmori T, Ohba M, et al. Distinct Afatinib Resistance Mechanisms Identified in Lung Adenocarcinoma Harboring an EGFR Mutation. Mol Cancer Res 2017;15:915-28. [Crossref] [PubMed]
- Chung JH, Rho JK, Xu X, et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer 2011;73:176-82. [Crossref] [PubMed]
- Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011;6:1152-61. [Crossref] [PubMed]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. [Crossref] [PubMed]
- Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016;1:e87062. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017.JCO2016719096. [PubMed]
- Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer 2007;58:411-3. [Crossref] [PubMed]
- McGranahan T, Nagpal S. A Neuro-oncologist's Perspective on Management of Brain Metastases in Patients with EGFR Mutant Non-small Cell Lung Cancer. Curr Treat Options Oncol 2017;18:22. [Crossref] [PubMed]
- Zeng YD, Liao H, Qin T, et al. Blood-brain barrier permeability of gefitinib in patients with brain metastases from non-small-cell lung cancer before and during whole brain radiation therapy. Oncotarget 2015;6:8366-76. [Crossref] [PubMed]
- Togashi Y, Masago K, Fukudo M, et al. Cerebrospinal fluid concentration of erlotinib and its active metabolite OSI-420 in patients with central nervous system metastases of non-small cell lung cancer. J Thorac Oncol 2010;5:950-5. [Crossref] [PubMed]
- Ahn MJ, Kim DW, Kim TM, et al. Phase I study of AZD3759, a CNS penetrable EGFR inhibitor, for the treatment of non-small-cell lung cancer (NSCLC) with brain metastasis (BM) and leptomeningeeal metastasis (LM). J Clin Oncol 2016;34:Abstr 9003.
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Niederst MJ, Hu H, Mulvey HE, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res 2015;21:3924-33. [Crossref] [PubMed]
- Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129-32. [Crossref] [PubMed]
- Uchibori K, Inase N, Araki M, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun 2017;8:14768. [Crossref] [PubMed]
- Scagliotti G, von Pawel J, Novello S, et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol 2015;33:2667-74. [Crossref] [PubMed]
- Gainor JF, Niederst MJ, Lennerz JK, et al. Dramatic Response to Combination Erlotinib and Crizotinib in a Patient with Advanced, EGFR-Mutant Lung Cancer Harboring De Novo MET Amplification. J Thorac Oncol 2016;11:e83-5. [Crossref] [PubMed]
- Dietrich MF, Yan SX, Schiller JH. Response to Crizotinib/Erlotinib Combination in a Patient with a Primary EGFR-Mutant Adenocarcinoma and a Primary c-met-Amplified Adenocarcinoma of the Lung. J Thorac Oncol 2015;10:e23-5. [Crossref] [PubMed]
- Ou SI, Govindan R, Eaton KD, et al. Phase I Results from a Study of Crizotinib in Combination with Erlotinib in Patients with Advanced Nonsquamous Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:145-51. [Crossref] [PubMed]
- Wu YL, Yang JC, Kim DW, et al. Safety and efficacy of INC280 in combination with geftinib (gef) in patients with EGFR-mutated (mut), MET-positive NSCLC: A single arm phase Ib/II study. J Clin Oncol 2014;35:abstr 8017.
- Wu YL, Soo RA, Kim DW, et al. Tolerability, efficacy and recommended phase II dose (RP2D) of tepotinib plus geftinib in Asian patients with c-Mer-positive/EGFR-mutant NSCLC: Phase Ib data. J Clin Oncol 2016;34:abstr e20501.
- Oxnard GR, Ramalingam SS, Ahn MJ, et al. Preliminary results of TATTON, a multi-arm phase Ib trial of AZD9291 combined with MEDI4736, AZD6094 or selumetinib in EGFR-mutant lung cancer. J Clin Oncol 2015;33:abstr 2509.
- Borczuk A, Paucar D, Halmos B. Has MET met its match? Ann Transl Med 2016;4:97. [Crossref] [PubMed]
- Gelsomino F, Facchinetti F, Haspinger ER, et al. Targeting the MET gene for the treatment of non-small-cell lung cancer. Crit Rev Oncol Hematol 2014;89:284-99. [Crossref] [PubMed]
- Noonan SA, Berry L, Lu X, et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. J Thorac Oncol 2016;11:1293-304. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first-and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer discovery 2016;6:1118-33. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Wilson FH, Johannessen CM, Piccioni F, et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell 2015;27:397-408. [Crossref] [PubMed]
- Tanizaki J, Okamoto I, Okabe T, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res 2012;18:6219-26. [Crossref] [PubMed]
- Miyawaki M, Yasuda H, Tani T, et al. Overcoming EGFR Bypass Signal-Induced Acquired Resistance to ALK Tyrosine Kinase Inhibitors in ALK-Translocated Lung Cancer. Mol Cancer Res 2017;15:106-14. [Crossref] [PubMed]
- Ou SI, Young L, Schrock AB, et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol 2017;12:137-40. [Crossref] [PubMed]
- Ko B, He T, Gadgeel S, et al. MET/HGF pathway activation as a paradigm of resistance to targeted therapies. Ann Transl Med 2017;5:4. [Crossref] [PubMed]
- Drilon A, Cappuzzo F, Ou SH, et al. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014;346:1480-6. [Crossref] [PubMed]
- Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 2015;21:1038-47. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Martinelli E, Morgillo F, Troiani T, et al. Cancer resistance to therapies against the EGFR-RAS-RAF pathway: The role of MEK. Cancer Treat Rev 2017;53:61-9. [Crossref] [PubMed]
- Fujita S, Masago K, Katakami N, et al. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J Thorac Oncol 2016;11:e67-e72. [Crossref] [PubMed]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP–dependent transporters. Nature Reviews Cancer 2002;2:48-58. [Crossref] [PubMed]
- Katayama R, Sakashita T, Yanagitani N, et al. P-glycoprotein mediates ceritinib resistance in anaplastic lymphoma kinase-rearranged non-small cell lung cancer. EBioMedicine 2015;3:54-66. [Crossref] [PubMed]
- Kim DW, Mehra R, Tan DS, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 2016;17:452-63. [Crossref] [PubMed]
- Gadgeel SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 2014;15:1119-28. [Crossref] [PubMed]
- Ou SH, Ahn JS, De Petris L, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol 2016;34:661-8. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Camidge DR, Bazhenova L, Salgia R, et al. Safety and efficacy of brigatinib (AP26113) in advanced malignancies, including ALK+ non–small cell lung cancer (NSCLC). J Clin Oncol 2015;33:abstr 8062.
- Kim DW, Tiseo M, Ahn MJ, et al. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol 2017.JCO2016715904. [PubMed]
- Qiao H, Lovly CM. Cracking the Code of Resistance across Multiple Lines of ALK Inhibitor Therapy in Lung Cancer. Cancer Discov 2016;6:1084-6. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015;33:992-9. [Crossref]
- Song A, Kim TM, Kim DW, et al. Molecular Changes Associated with Acquired Resistance to Crizotinib in ROS1-Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:2379-87. [Crossref] [PubMed]
- Zou HY, Li Q, Engstrom LD, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A 2015;112:3493-8. [Crossref] [PubMed]
- Drilon A, Somwar R, Wagner JP, et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin Cancer Res 2016;22:2351-8. [Crossref] [PubMed]
- Dziadziuszko R, Le AT, Wrona A, et al. An Activating KIT Mutation Induces Crizotinib Resistance in ROS1-Positive Lung Cancer. J Thorac Oncol 2016;11:1273-81. [Crossref] [PubMed]
- Cargnelutti M, Corso S, Pergolizzi M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget 2015;6:5182-94. [Crossref] [PubMed]
- Davies KD, Mahale S, Astling DP, et al. Resistance to ROS1 inhibition mediated by EGFR pathway activation in non-small cell lung cancer. PLoS One 2013;8:e82236. [Crossref] [PubMed]
- Rossing HH, Grauslund M, Urbanska EM, et al. Concomitant occurrence of EGFR (epidermal growth factor receptor) and KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) mutations in an ALK (anaplastic lymphoma kinase)-positive lung adenocarcinoma patient with acquired resistance to crizotinib: a case report. BMC Res Notes 2013;6:489. [Crossref] [PubMed]
- Gimenez-Xavier P, Pros E, Bonastre E, et al. Genomic and molecular screenings identify different mechanisms for acquired resistance to MET inhibitors in lung cancer cells. Mol Cancer Ther 2017;16:1366-76. [Crossref] [PubMed]
- Li A, Yang J, Zhang XC, et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non-small cell lung cancer. Clin Cancer Res 2017;23:4929-37. [Crossref] [PubMed]
- Cepero V, Sierra JR, Corso S, et al. MET and KRAS gene amplification mediates acquired resistance to MET tyrosine kinase inhibitors. Cancer Res 2010;70:7580-90. [Crossref] [PubMed]
- McDermott U, Pusapati RV, Christensen JG, et al. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res 2010;70:1625-34. [Crossref] [PubMed]
- Lee NV, Lira ME, Pavlicek A, et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells through [corrected] MAPK activation. PLoS One 2012;7:e39653. [Crossref] [PubMed]
- de Langen AJ, Smit EF. Therapeutic approach to treating patients with BRAF-mutant lung cancer: latest evidence and clinical implications. Ther Adv Med Oncol 2017;9:46-58. [Crossref] [PubMed]
- Lin L, Asthana S, Chan E, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci U S A 2014;111:E748-57. [Crossref] [PubMed]
- Yao Z, Torres NM, Tao A, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell 2015;28:370-83. [Crossref] [PubMed]
- Johnson DB, Menzies AM, Zimmer L, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer 2015;51:2792-9. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Levy B, Hu ZI, Cordova KN, et al. Clinical Utility of Liquid Diagnostic Platforms in Non-Small Cell Lung Cancer. Oncologist 2016;21:1121-30. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Ihuegbu N, Banks KC, Fairclough SR, et al. Non-invasive detection of crizotinib resistance in ALK-rearranged lung adenocarcinoma directs treatment with next-generation ALK inhibitors. J Clin Oncol 2016;24:abstr e20643.
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 2016;7:10501. [Crossref] [PubMed]
- Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol 2015;33:1152-8. [Crossref] [PubMed]
- Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819-29. [Crossref] [PubMed]
- Anagnostou V, Smith KN, Forde PM, et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:264-76. [Crossref] [PubMed]