In vitro prediction of breast cancer therapy toxicity
Introduction
Approximately 50% of cancer patients receive radiotherapy (RT) (1) with sub-population (~1–5%) developing significant normal tissue complications within the treatment field, limiting safe dose-escalation across the general RT population. Various assays have been trialled in an attempt to predict excessive normal tissue toxicity prior to the instigation of therapy, with no practical assay useful in the clinical setting. In this study we characterize a unique bank of cell samples from radiosensitive (RS) patients, utilizing an assay that allows for cross-comparison with patient characteristics, to investigate its predictive clinical outcome power.
Variation in normal tissue reactions in the cancer patient population has been observed as being normally distributed, ranging from RS to radioresistant sub-populations, to extreme over-reactors lying outside the normally distributed curve (2). For a large group of patients treated with the same techniques, a small number show a highly sensitive normal tissue response, with either severe acute or severe late radiation reactions. In some cases, severe acute reactions progress to severe late effects, so-called “consequential late effects”. Some variability among individuals in normal tissue response can be explained by Poisson randomness in cell killing (2), with a genetic basis for radiosensitivity suggested by studies using cells from individuals with a RS phenotype, such as ataxia-telangiectasia (AT) and Nijmegen breakage syndrome (NBS) (3,4). Such individuals are readily identified by their abnormal clinical characteristics, including in addition to radiosensitivity, a Mendelian pattern of genetic transmission, immunodeficiency and cancer-proneness, especially lymphoid malignancies. In contrast to such patients, the present study was limited to patients with no additional discernible phenotypes apart from clinical radiosensitivity.
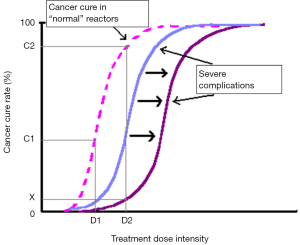
In conjunction with predicting inherent radiosensitivity and hence relieving treatment morbidity, identification of individuals with sensitive normal tissues using a predictive assay could theoretically allow dose escalation in the majority of patients, with the aim of increasing local control and cure rates (5-7) (Figure 1). In spite of enormous attempts to develop a predictive assay recapitulating intrinsic clinical radiosensitivity, there is still conflicting evidence regarding the correlation of in vitro radiosensitivity and in vivo responses to IR, especially in breast cancer (8); the field requires further investigation.
Tissues regenerate clonally after irradiation, for example, in the skin (9). Colony-forming assays using fibroblasts are regarded by many as the gold-standard measurement of in vitro clonal cell survival (10). Alternative clonogenic assays such as the limiting dilution assay (LDA), as used here and which are suitable for non-adherent lymphoid cells, are based on the same principle as the CFA in its ability to assess clonal survival of a cell population after radiation exposure in vitro. Whatever assay is used to measure in vitro radiosensitivity, all studies have found a great deal of inter-individual variability. Clinical observation of IR-induced normal tissue damage supports the notion that intrinsic differences in late radiation injury to normal tissues may have a genetic basis (11-13).
Directly demonstrating a genetic factor contributing to non-syndromic human radiosensitivity, a large genome-wide association study with requisite power demonstrated a novel candidate radiosensitivity gene, TANC1, within a defined genomic region in a RS prostate cancer cohort compared to control (14). Mutations in this gene were associated with late RT toxicity. The gene had previously been linked to neurological phenotypes and in regenerating damaged muscle, the latter compatible with a role in late IR toxicity. The gene is thus a potential marker for RT side-effects in the prostate cancer RT patients of the type studied in this analysis.
Other studies have attempted to identify molecular markers of clinical radiosensitivity (15,16). These include using a candidate gene/protein approach and studying molecules known or suspected to influence radiosensitivity in other species, but using IR-sensitive patient cells/tissues. There are a number of examples of such studies (17-21). Increasingly, gene expression profiling is giving insights into human disease and such studies are emerging in the field of radiosensitivity (22-24).
If there is a general genetic basis for the observed differences in clinical radiosensitivity in otherwise phenotypically normal cancer patients, then it is reasonable to expect a relationship between radiosensitivity of different normal tissues within an individual and some differences in in vitro IR-responses between normal and sensitive patients examined. However, evidence to date does not allow such a conclusion.
Using lymphoblastoid cell lines (LCLs) from patients with the LDA has some advantages over other cellular survival assays. LCLs are easily derived from blood samples, which can be collected from patients with minimal pain and discomfort compared to skin biopsies. Such cells are immortalized, therefore providing a robust source of cells available for use in different assays. The LDA can easily provide many replicates, increasing statistical power.
The aim of this study was to examine normal tissue responses to IR using the LDA on immortalized peripheral B-lymphocytes (LCLs) as a model, with the ultimate and main goal of assessing its suitability as a predictive test for clinical application in RT. The data obtained was related to clinicopathological manifestations of a unique RS patient cohort and was compared with pooled controls. If the severe normal tissue reactions have a genetic basis, clinically sensitive individuals may have radiosensitivity that is distinguishable from the control cohort.
Methods
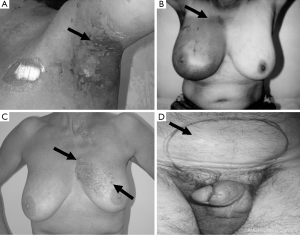
Ethics approval (96/39) for this study, employing cell lines from cancer patients, was obtained from the Ethics Committee of the Peter MacCallum Cancer Centre, Melbourne, Australia. Patients gave individual consent for the creation of LCLs and for their use in research. A group of 29 RT patients treated between 1987 and 1999 were recruited for this study. Patients with Radiation Therapy Oncology Group (RTOG) grade 3 or 4 toxicities (Figure 2) (one case had grade 2 toxicity) were designated as being clinically RS, with those with RTOG scores ≤1 serving as controls. The study cohort consisted of LCLs from 9 patients with severe acute reactions, 4 with consequential late reactions, 6 with severe late reactions only, 10 controls and 1 AT patient. Most controls were matched for a number of factors, including age, sex, ethnicity, cancer type and total radiation dose, with corresponding RS patient cells. The overall group represented 5 cancer subtypes, with a predominance of breast cancer among the radiosensitivity patients (n=14). All patients selected for this study were treated with a fractionated RT schedule using 6 MV photons, 2 Gray (Gy) per fraction, apart from one prostate cancer patient who received 18 MV photons. Total dose and duration varied depending on cancer type and occurrence of acute radiation reactions. Mean date of therapy completion for acute and consequential late reaction patients was 1997, while the mean completion date for patients with late reactions was 1993.
Blood samples were spun down for lymphocyte isolation; B-cells immortalized with Epstein-Barr virus (EBV), expanded in culture and cryo-preserved in liquid nitrogen storage tanks. LCLs were also obtained from an AT patient to serve as an IR sensitive control.
For the LDA assay, LCLs were irradiated at room temperature as a cell suspension with γ-radiation from a 137Cs source, delivered at 1 Gy/1.497 min. Cells from the AT patient were irradiated at 1, 1.5, and 2 Gy, due to the extreme radiosensitivity of AT cell lines, with the other cell lines irradiated at 1, 2, and 3 Gy.
For the LDA, cells were fed 24 hours prior to the assay. A cell count was carried out on cell suspensions using the Sysmex (Bayswater, Victoria, Australia) counter system. Only cell lines with more than 3×105 cells/mL were used for experimentation. Cell suspensions were then diluted to 10–15× the final plating cell concentration and 4 mL of the suspension was used for each irradiation dose (Figure 3A). An aliquot of 4 mL was mock irradiated (0 Gy) at room temperature. This served as a baseline for cell survival before radiation exposure (100% survival). Samples at each dose level were serially diluted to four concentrations, and 160 µL/well put into 96-well, round-bottomed plates. The outside wells of each plate were filled with 200 µL of phosphate-buffered saline to reduce evaporation from the inside wells. For a preliminary test for each cell line, six different concentrations, each in 10 wells, were used to determine the four most appropriate cell concentrations for plating (Figure 3A). After pilot studies were carried out, each of the four concentrations was plated into 20 wells in triplicate. The full experiment was repeated at least in duplicate on a separate occasion to ensure accuracy and reproducibility.
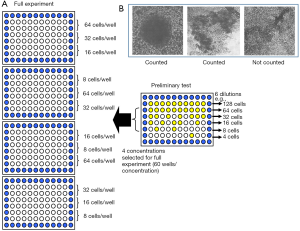
Plates were incubated at 37 °C in humidified air with 5% CO2 for 2 weeks without a medium change. At day 14, 40 µL of a 0.5 mg/mL solution of MTT dissolved in PBS was added to each well, then plates were reincubated for 2–4 hours to allow the mitochondrial enzymes to reduce the MTT to an insoluble dark blue formazan salt (25). Wells with dark blue cellular aggregates containing more than 50 cells were scored as a colony (Figure 3B).
LDA analysis was performed on six of 30 LCLs using 10% human serum in place of 10% FBS. These cell lines were RS10 (an AT line), CL20, CL29, RS9, RS29 and RS127. There is some evidence that human serum gives higher cloning efficiency than FBS (10). However, with repeat experiments on these cell lines using FBS as serum no significant difference in cell survival was seen. Therefore, media supplemented with 10% FBS was subsequently used throughout the study. The effect of FBS itself on cell growth may vary from batch to batch; therefore the same batch of FBS was used for all experiments to minimize this variation.
Most samples were coded and blinded, including RS6, RS12, RS18, RS21, RS28, RS43, RS106, RS112, CL7, CL21, CL22, CL32, CL38, CL39, CL44 and CL103. This protocol was suggested after a number of LDAs had been performed on some cell lines, with the aim of reducing bias in data collection.
Results
The LDA, a surrogate for clonogenic survival, was performed on a total of 30 LCLs in at least two repeat assays on separate occasions except for two cell lines, RS28 and RS38, which were tested just once. The mean SFs at all irradiation doses were obtained from each of 28 LCLs in replicate, and single values of SF from the other two LCLs. Thirty survival curves were generated using these values.
LCL radiosensitivity was determined as both Gray [the surviving fraction at 2 Gray (SF2)] and D0 (the latter being the slope of cell survival curve), both obtained from the fitted survival curves. SF2 values for all cell lines ranged from 0.0245 to 0.3894 (mean 0.2521±0.0791 SD). The D0 values ranged from 0.539 to 2.071 Gy (mean 1.463±0.3727 SD), using Spearman’s rank correlation test (26). Both IR sensitivity parameters of cell survival equally well represented the cellular radiosensitivity of LCLs (P<0.0001). Thereafter the subsequent description of radiosensitivity was also expressed in terms of SF2.
A range of sensitivities was observed across LCLs exposed to graded doses of IR (Figure 4). The mean radiosensitivity of the breast cancer patients was greater than, and clearly distinguishable from, that of the controls. Both occupied their own distinctive zones on the cell survival curves. When the patients were grouped according to their cancer types, control breast cancer patients (n=8) had a higher mean SF2 than breast cancer patients who were clinically sensitive to RT (n=14) (P=0.011). Other cancer types did not have enough controls for statistical analysis. However, SF2 appeared to be related to the cancer type, with prostate cancer patients having more resistant LCLs and head and neck cancer patient LCLs being the most in vitro sensitive. However, the significance of these differences could not be evaluated because of an inadequate number of samples.
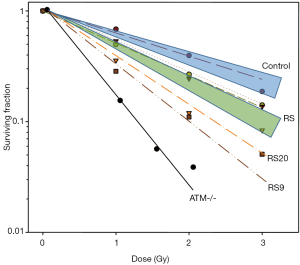
There were also two notable patient RS strains, RS20 and RS9, from patients with breast cancer and head and neck cancer, respectively. When comparing the difference of the outliers and the mean of all controls, RS20 was marginally more sensitive (P=0.0682), whilst RS9 was significantly more sensitive than the controls (P=0.0329). None of the samples equalled the RS control line, the AT homozygote, in sensitivity as may have been expected.
The coefficient of variation (CV) for SF2 (the ratio of standard error and the mean, that indicates the degree of variation among parameters in question, in this case, SF2 observed in two experiments per cell line) for the whole group of 30 cell lines was 31%, while the CV for D0 was 22%. Figure 5 demonstrates the SF2 variation between different cell lines. Variation between individuals was greater than the variation between experiments for most cell lines, with the most dramatic example being CL20, whose SF2 in one experiment was the most radioresistant but which was quite RS in a repeat experiment. The radiosensitivity variation in control cell lines, on average, was greater than the radiosensitivity lines.
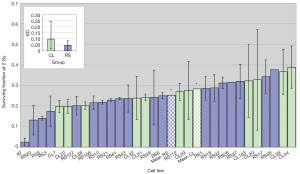
The role of patients’ age, cancer type and total dose received in influencing in vitro radiosensitivity was examined. The mean age of patients when the samples were collected (age at sampling) was 59.72 years old, and ranged from 32–82 years. The age at sampling did not influence their LCL radiosensitivity (r=0.0019; P=0.9927). Five cancer types were included in this study with most having breast cancer. Regarding RT schedules, most breast cancer patients received 50 Gy in total, compared to 66 Gy for 2 of 3 prostate cancer patients. The lowest total dose was 16.5 Gy given to a patient with seminoma. Correlation between total dose and SF2 was not significant (r=0.02759; P=0.8913).
Discussion
Using the LDA, this study investigated the cellular response to IR of LCLs from cancer patients who had or had not experienced severe clinical reactions to RT. Our aim was to determine whether cells from clinically RS patients also manifested in vitro IR sensitivity. The rationale was 2-fold: firstly to identify any case of in vitro radiosensitivity for further characterization (and potentially to shed light on the cellular and molecular basis of radiosensitivity) and secondly to determine whether the assay could be viable as a predictive assay for radiosensitivity in the clinic. Clinical radiosensitivity is a major issue for the 50% of cancer patients who will undergo RT. For those who suffer severe acute or late toxicities, the morbidity is significant and in some cases debilitating. Fortunately, such reactions are uncommon. Predicting such reactions to IR prior to treatment based on in vitro responses could allow improvement of the therapeutic ratio in RT and may have application in a clinical setting to guide therapy, for example, by allowing IR dose de-escalation (as has been used previously in highly IR-sensitive AT patients) or the use of cancer therapies other than radiation.
We created cell survival curves using non-linear regression analysis. Comparison with SF2 values showed that the latter could substitute for the former, simplifying future use of this assay for LCLs.
Across all tested samples, we found a statistical trend towards separation of radiosensitivity and control cell survival curves. More importantly, we found that the LDA could distinguish between controls and LCLs from clinically RS cases of breast cancer. This suggests that the LDA may provide unique clinical utility as a predictive assay in breast cancer, especially when patients were undergoing neoadjuvant or adjuvant chemotherapy, when LCLs could be created and LDA-tested during the period of chemotherapy. Such data could potentially facilitation of IR schedules based on the LDA results, or patients could be treated with alternative therapeutic modalities. The number of cases in this study could be criticized based on small sample size. However, such severe RS cases are rare, and the statistics we used, robust. These data should nevertheless be confirmed in larger studies with prospectively-accessioned cases, although the relative rarity of severely RS cases may impede this approach.
The utility of the LDA in cases of hereditary breast cancer, where survival curves after radiation exposure failed to distinguish controls from BRCA1 or BRCA2 carriers (27), has yet to be determined.
In this study two distinctively in vitro IR sensitive cases were also found. The lymphocytic cells from these two patients are clearly of a different type from their corresponding cells and tissues showing clinical radiosensitivity, suggesting that IR sensitivity in these cases was constitutive, although tissue-specific differences in radiosensitivity of mammals are described (28). These cell lines may be useful for further molecular and other studies of radiosensitivity.
In summary, our main finding was that the LDA distinguished control cell lines from breast cancer patient-derived lymphoblasts and may be a new predictive assay to allow personalization of RT in breast cancer.
Acknowledgements
Funding: Australian National Health and Medical Research Council Project Grant 145780 to MJ McKay.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee of the Peter MacCallum Cancer Centre (96/39) and written informed consent was obtained from all patients.
References
- Radiation Therapy for Cancer. Available online: https://www.cancer.gov/about-cancer/treatment/types/radiation-therapy/radiation-fact-sheet
- Burnet NG, Johansen J, Turesson I, et al. Describing patients' normal tissue reactions: concerning the possibility of individualising radiotherapy dose prescriptions based on potential predictive assays of normal tissue radiosensitivity. Steering Committee of the BioMed2 European Union Concerted Action Programme on the Development of Predictive Tests of Normal Tissue Response to Radiation Therapy. Int J Cancer 1998;79:606-13. [Crossref] [PubMed]
- Arlett CF, Harcourt SA. Survey of radiosensitivity in a variety of human cell strains. Cancer Res 1980;40:926-32. [PubMed]
- Cole J, Arlett CF, Green MH, et al. Comparative human cellular radiosensitivity: II. The survival following gamma-irradiation of unstimulated (G0) T-lymphocytes, T-lymphocyte lines, lymphoblastoid cell lines and fibroblasts from normal donors, from ataxia-telangiectasia patients and from ataxia-telangiectasia heterozygotes. Int J Radiat Biol 1988;54:929-43. [Crossref] [PubMed]
- Peters L, McKay M.. Predictive assays: will they ever have a role in the clinic? Int J Radiat Oncol Biol Phys 2001;49:501-4. [Crossref] [PubMed]
- Russell NS, Begg AC. Editorial radiotherapy and oncology 2002: predictive assays for normal tissue damage. Radiother Oncol 2002;64:125-9. [Crossref] [PubMed]
- Tucker SL, Geara FB, Peters LJ, et al. How much could the radiotherapy dose be altered for individual patients based on a predictive assay of normal-tissue radiosensitivity? Radiother Oncol 1996;38:103-13. [Crossref] [PubMed]
- Loeffler JS, Harris JR, Dahlberg WK, et al. In vitro radiosensitivity of human diploid fibroblasts derived from women with unusually sensitive clinical responses to definitive radiation therapy for breast cancer. Radiat Res 1990;121:227-31. [Crossref] [PubMed]
- McKay MJ, McKay C. Clonal wound re-epithelialisation after radiotherapy for skin cancer. Australas J Dermatol 2015;56:e91-2. [Crossref] [PubMed]
- Begg AC, Russell NS, Knaken H, et al. Lack of correlation of human fibroblast radiosensitivity in vitro with early skin reactions in patients undergoing radiotherapy. Int J Radiat Biol 1993;64:393-405. [Crossref] [PubMed]
- Bentzen SM, Overgaard M, Overgaard J. Clinical correlations between late normal tissue endpoints after radiotherapy: implications for predictive assays of radiosensitivity. Eur J Cancer 1993;29A:1373-6. [Crossref] [PubMed]
- Geara FB, Peters LJ, Ang KK, et al. Prospective comparison of in vitro normal cell radiosensitivity and normal tissue reactions in radiotherapy patients. Int J Radiat Oncol Biol Phys 1993;27:1173-9. [Crossref] [PubMed]
- Sprung CN, Mathews LJ, McKay MJ. DNA repair deficiencies: connecting carcinogenesis and sensitivity to ionising radiation. Today's Life Science 2002;14:40-4.
- Fachal L, Gómez-Caamaño A, Barnett GC, et al. A three-stage genome-wide association study identifies a susceptibility locus for late radiotherapy toxicity at 2q24.1. Nat Genet 2014;46:891-4. [Crossref] [PubMed]
- West CM, McKay MJ, Hölscher T, et al. Molecular markers predicting radiotherapy response: report and recommendations from an International Atomic Energy Agency technical meeting. Int J Radiat Oncol Biol Phys 2005;62:1264-73. [Crossref] [PubMed]
- Kerns SL, West CM, Andreassen CN, et al. Radiogenomics: the search for genetic predictors of radiotherapy response. Future Oncol 2014;10:2391-406. [Crossref] [PubMed]
- Fang Z, Kozlov S, McKay MJ, et al. Low levels of ATM in breast cancer patients with clinical radiosensitivity. Genome Integr 2010;1:9. [Crossref] [PubMed]
- Severin DM, Leong T, Cassidy B, et al. Novel DNA sequence variants in the hHR21 DNA repair gene in radiosensitive cancer patients. Int J Radiat Oncol Biol Phys 2001;50:1323-31. [Crossref] [PubMed]
- Leong T, Whitty J, Keilar M, et al. Mutation analysis of BRCA1 and BRCA2 cancer predisposition genes in radiation hypersensitive cancer patients. Int J Radiat Oncol Biol Phys 2000;48:959-65. [Crossref] [PubMed]
- Fogarty GB, Muddle R, Sprung CN, et al. Unexpectedly severe acute radiotherapy side effects are associated with single nucleotide polymorphisms of the melanocortin-1 receptor. Int J Radiat Oncol Biol Phys 2010;77:1486-92. [Crossref] [PubMed]
- Vasireddy RS, Sprung CN, Cempaka NL, et al. H2AX phosphorylation screen of cells from radiosensitive cancer patients reveals a novel DNA double-strand break repair cellular phenotype. Br J Cancer 2010;102:1511-8. [Crossref] [PubMed]
- Alsner J, Rødningen OK, Overgaard J. Differential gene expression before and after ionizing radiation of subcutaneous fibroblasts identifies breast cancer patients resistant to radiation-induced fibrosis. Radiother Oncol 2007;83:261-6. [Crossref] [PubMed]
- Forrester HB, Ivashkevich A, McKay MJ, et al. Follistatin is induced by ionizing radiation and potentially predictive of radiosensitivity in radiation-induced fibrosis patient derived fibroblasts. PLoS One 2013;8:e77119. [Crossref] [PubMed]
- Sprung CN, Li J, Hovan D, et al. Alternative transcript initiation and splicing as a response to DNA damage. PLoS One 2011;6:e25758. [Crossref] [PubMed]
- Hayon T, Dvilansky A, Shpilberg O, et al. Appraisal of the MTT-based assay as a useful tool for predicting drug chemosensitivity in leukemia. Leuk Lymphoma 2003;44:1957-62. [Crossref] [PubMed]
- Myers JL, Well AD. Research Design and Statistical Analysis. 2nd edition. USA: Lawrence Erlbaum, 2003:508.
- Lovelock PK, Wong EM, Sprung CN, et al. Prediction of BRCA1 and BRCA2 mutation status using post-irradiation assays of lymphoblastoid cell lines is compromised by inter-cell-line phenotypic variability. Breast Cancer Res Treat 2007;104:257-66. [Crossref] [PubMed]
- Hendry JH, Jiang TN. Differential radiosensitising effect of the scid mutation among tissues, studied using high and low dose rates: implications for prognostic indicators in radiotherapy. Radiother Oncol 1994;33:209-16. [Crossref] [PubMed]


