The mycobiome of the human urinary tract: potential roles for fungi in urology
Introduction
The microbiome, the aggregate collection of microorganisms within a physiologic niche, modulates human physiology in ways that we are just beginning to comprehend, influencing organ permeability and barrier function, vitamin synthesis, metabolism, neurological activity, and inflammation and immunity (1,2). Studies of these microbial communities were previously hindered by limitations of culture methods, but the development of culture-independent discovery techniques aimed at the detection of microbial DNA has revealed complex populations in sites previously thought to be sterile, such as the lung, brain, and breast. This insight prompted multiple comprehensive studies looking at bacterial ecosystems and their role in health and disease over the past decade, exemplified by the large-scale National Institutes of Health (NIH) Human Microbiome Project (HMP) in the United States and the Metagenomics of the Human Intestinal Tract (MetaHIT) Project in Europe.
Before the advent of culture-independent analyses, the uninfected urinary tract had been assumed to be a sterile environment and was therefore not included in the HMP (3). It has since become clear that bacterial communities exist within the urinary tract, even in healthy, asymptomatic subjects (4-8). Large shifts in urinary bacterial communities are found in a variety of pathologies, such as neurogenic bladder, stress and urgency incontinence, interstitial cystitis/painful bladder syndrome (IC/PBS), and sexually transmitted infections (4-7,9-13), suggesting the resident microbial communities change in disease states [reviewed in (14)].
With the development of affordable, rapid, and scalable culture-independent methods for the study of bacterial communities, the last decade has seen a massive expansion in studies aimed at profiling commensal bacterial communities. Despite this new interest in and understanding of the importance of microbial communities in human physiology, non-bacterial populations, such as fungi, viruses, archae, and protozoa, have remained understudied. As the term “microbiome” is frequently used to refer to the study of bacteria alone, the term “mycobiome”, a combination of microbiome and mycology, emerged in 2010 (15) to designate the endogenous community of fungi within a specific host anatomic location. This field is still in its infancy; the term “mycobiome” or “fungal microbiome” only appears in PubMed-indexed articles 105 times. Of these articles, only 37 provide original data regarding communities in human hosts, several of which utilize only culture-based methods. None of these examines the urinary tract.
There are several explanations for the inattention to the role of fungi in host physiology, including a lack of standardized information regarding the vast numbers of species, a lack of reagents and tools for the study of fungal populations, and a (potentially) erroneous belief that these organisms are not as critical as bacteria in human disease. Only a few fungal species are known to cause invasive infections in humans despite hundreds of thousands of fungal species throughout the environment (16). The pathogenic potential of these organisms is clear; while rare, fungal infections of the urinary tract can be life-threatening due to the limitations in culture methodologies, frequent low suspicion for fungal involvement, and the lack of preventative and therapeutic options (17). Candida spp. are overwhelmingly the most prevalent urinary pathogen, but all of the common invasive fungal species, such as Cryptococcus, Aspergillus, Mucoraceae, Histoplasma, Blastomyces, and Coccidioides, can infect the urinary tract. Recent epidemiological studies have noted increases in both the populations infected and in the development of more virulent organisms resistant to common antifungals (18,19). This shift may be due to increases in the use of antineoplastic drugs, systemic immunosuppressants, prosthetic grafts and implants, as well as the wide-spread use of broad-spectrum antibiotics (20).
It is becoming clear that there are associations between changes in the mycobiome and common, significant diseases in the immunocompetent host such as hepatitis, cystic fibrosis, and inflammatory bowel disease (IBD), which can be seen in the absence of frank, fungal infections. But is this the case for fungi in the urinary tract? As early as the 1850s, clinicians were able to demonstrate viable fungi in the urine of asymptomatic subjects and note alterations in fungal composition in association with diabetes and renal disease, again, in the absence of overt infection (21). But more than 150 years have gone by with little increase in our knowledge or understanding of urinary fungi. A substantive portion of this lack of progress may be due to the lack of tools available to identify and study this kingdom and its interactions with the human host.
Tools to study the human mycobiome
Culture techniques
The majority of fungal culture techniques date back to the 1920s with few changes in the past 100 years. Identifications are made by examining growth structures and evaluating growth on different media (22). Even for cultivatable species, low abundance organisms and organisms that require microbe-microbe interactions cannot be cultivated optimally (23). More importantly, unculturable fungi comprise the largest part of the human mycobiome. In studies of the pulmonary fungal and bacterial microbiome in cystitc fibrosis, 60% of genera were missed by culture (24). In one study of the oral mycobiome, 11 of 85 fungal genera identified by culture-independent methods could not be cultured at all (15). In 30% of the participants, non-cultivatable fungi were 50% or more of the fungi identified. In a direct comparison, extended culture conditions identified five species, while culture-independent analyses identified 37 unique fungal genera within the gut mycobiome (25). Although culture will remain an important tool in our armamentarium, culture-based methods are a poor methodology to examine the complexity and depth of commensal fungal communities.
Culture-independent methods
At the heart of culture-independent approaches to the detection of microorganisms is an assumption that DNA isolated from an anatomic site will contain genomic sequences from resident microbes which provide a snapshot of the ecosystem at that moment in time. Several methodologies allow the rapid assessment of the relative diversities of samples without a need for specialized expertise and expensive equipment, such as restriction fragment length polymorphism (RFLP) analysis, oligonucleotide fingerprinting of rRNA genes (OFRG), denaturing gradient gel electrophoresis (DGGE) and in situ hybridization. While these methods can be informative, they lack the specificity to identify fungal differences in larger populations at the species level.
In general, direct sequencing methods provide the best representation of diversity and complexity in microbial community profiling. Shotgun metagenomics and metatranscriptomics, which involve the sequencing and analysis of total genomic DNA or RNA transcripts respectively (26,27), theoretically provide the most comprehensive and unbiased data, but have proven challenging in the analysis of fungi from human biological samples given their low abundance. Previous attempts in other organ systems have been unable to produce data sets with enough sequence data to reconstruct the genomes of fungi from complex human samples such as stool (26).
The majority of mycobiome studies utilize next generation sequencing (NGS) of pools of specific DNA targets. After isolation of DNA from a biological sample, small variable regions of the fungal genome are amplified by polymerase chain reaction (PCR), followed by large-scale pyrosequencing of thousands or even millions of these fragments. The taxon corresponding to each fragment is identified by structured comparison to a database. In comparison to traditional Sanger sequencing, NGS has made community-level analyses more efficient and cost-effective, making it the effective gold standard for microbiome profiling. There are, however, significant limitations to this technology and unique challenges for fungal identification at every step of sample processing and analysis.
Challenges in fungal community profiling
DNA extraction
Previous studies examining the optimization of DNA isolation and purification for analysis of bacterial species (“microbiome”) in samples with a standardized, diverse bacterial community demonstrated that different sample preparation and DNA extraction protocols have dramatically different results (1,15,28,29). While more extensive protocols utilizing mechanical disruption by bead beating and multiple enzymatic treatments consistently gave the best representations of bacterial community structure, in no case did the obtained results provide a completely accurate picture of standardized samples (29). Isolation of DNA from fungi can be even more challenging than from bacterial cells, as fungi are often more structurally robust and difficult to lyse effectively. As no extraction method will disrupt all species uniformly, hardier fungi may be underrepresented in analyses. In addition, the optimal cellular disruption/lysis and subsequent DNA extraction and purification conditions for fungal samples have not been clearly defined (30). These conditions may differ significantly from those used for bacterial isolation and may influence both overall DNA yields and relative species representation.
Amplification methodology
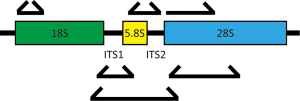
In contrast to metagenomics, methods based on sequencing pools of specific DNA targets typically rely on the selection of a relatively conserved genomic region to serve as a proxy for the entire genome. As in bacterial profiling, these sequences are amplified using primers designed to hybridize to regions conserved between fungi but containing between them areas of sufficient variability to allow taxonomic assignment. For profiling of bacteria, the 16S rRNA locus is targeted. The fungal rDNA cluster of genes encodes the 18S, 5.8S, and 28S ribosomal subunits in a single locus separated by two internal transcribed spacers (ITS1 and ITS2) (Figure 1). Multiple regions within this locus have been utilized for community profiling, most commonly the 28S large subunit (LSU), 18S small subunit (SSU) and the ITS. The LSU is highly conserved but lacks discriminatory power for many species. The SSU is less conserved than the LSU, providing better phylogenetic distinction, and allows the detection of a number of non-fungal eukaryotes, such as parasitic protozoa (28,31). The ITS regions are most commonly used as the higher diversity at these loci enable greater genus-level phylogenetic assignment (32). This diversity, however, can lead to underamplification of certain families, inevitably biasing the results towards certain species. For example, commonly-used ITS1 primer combinations can be biased toward amplification of Basidiomycetes, while ITS2 primer sets can be biased towards the Ascomycetes (33,34).
Sample processing and analysis
The sequencing platform utilized can also impact the quality and character of information obtained by NGS (30,35). Pyrosequencing using the 454 GS-FLX (Roche Diagnostics Corporation) technology is the most expensive, but produces the longest sequence reads (>500 bp) (36). The Illumina© HiSeq and MiSeq platforms (Illumina, Inc.) dominate the NGS market, providing a good balance of convenience and quality. HiSeq gives the highest data output at lowest cost, while MiSeq is better balanced for quick turnaround time and longer read lengths (37,38). The Ion Torrent systems (Thermo Fisher Scientific, Inc.) offer low cost, scalable, high-throughput sequencing competitive with the Illumina technology (39), although we have noted that it has more trouble handling the diverse amplicon lengths generated when amplifying ITS1 or ITS2 regions (30).
Once sequencing is complete, pipelines for the analysis of sequence data compile bioinformatics tools that allow the trimming, screening, and alignment of sequences for the assignment of operational taxonomic units (OTUs), phylogenetic analyses, and determinations of fungal diversity within and across groups (α and a diversity). The QIIME (40) and Mothur (41) analytical methods are widely used for all varieties of microbial profiling. More recent tools, such as CloVR-ITS (32) and BROCC (28), were uniquely developed for mycobiome studies, using customized automated analysis pipelines containing elements from both QIIME and Mothur to optimize taxonomic assignment of ITS and 18S amplicons, respectively.
Taxonomic classification
The most limiting step in mycobiome analyses is the choice of sequence database. There does not exist for fungi a database as rich as that available for bacterial 16S rDNA. For ITS sequences, the UNITE database (42) is most commonly used and contains the greatest number of overall annotated fungal sequences, but may not be best for the analysis of human biological samples. The targeted host-associated fungi (THF) database, designed specifically for biologic, not environmental, fungal discovery, consistently outperforms UNITE and other large ITS1 databases (30) on comprehensiveness, taxonomy assignment accuracy, and computational efficiency in analyzing sequencing data from mammalian sources. There exist a multitude of additional sequence collections, such as the SILVA database of 18S and 28S sequences (43) and the PHYMYCO-DB collection of SSU and EF1-α gene sequences (44).
While the curation of public databases has significantly improved in the past decade, a 2006 estimate suggested that only 1% of fungal species were represented in GenBank and that approximately 20% of entries may be misclassified to the species level (45). Also lacking from these databases is the capability to categorize fungi at the subspecies level. Additional problems arise for fungi which have not been addressed or resolved in the literature given the lack of systematic nomenclature for fungi; e.g., a single species may have been given multiple names. In addition, sexual and asexual forms of the same fungal species are often classified as different taxa (28). These resources are themselves evolving and improving, but require continued annotation and curation to move forward.
Relative abundance readouts
The ability of NGS technology to amplify and detect very low abundance species provides tremendous analytic power, but this sensitivity is also a source of significant consternation; contamination and context can influence results, particularly in low density samples. Quantification of absolute fungal levels is not possible with NGS alone; these analyses typically only provide a relative abundance of detection of specific sequences within the overall population. Trace environmental contamination, carryover of populations from nearby sites, variations in sample quantity or quality, and even minor fluctuations in physiologic conditions may result in large, misleading population shifts. The predominance of a fungal species may represent robust expansion of that population, selective survival of that species under selective pressure, or both. While the significance of these situations is different, as are the absolute numbers of live fungi, there is no way to discriminate these possibilities based on NGS data alone. Further, rDNA loci in bacteria and fungi are generally present in genomes in multiple (and often variable) copy-numbers. Thus, it is critical to combine NGS-based determinations of microbial changes with alternative molecular and microbiological techniques to draw conclusions about the roles of particular population shifts in disease conditions.
Challenges in evaluating the urinary mycobiome in particular
Host contamination
As fungi are eukaryotes, the presence of non-fungal eukaryotic DNA (e.g., from host cells) can compete with fungal DNA for reagents during extraction and amplification. The more human material, the worse this interference becomes. Thus, contaminating human cells may interfere with the specific amplification and sequencing of fungal ribosomal sequences. In urinary samples, the ratio of fungi to sloughed human urothelial (and contaminating epithelial) cells is unclear, but presumed to be extremely low, reducing the sensitivity of mycobiome analyses in lower urinary tract disease.
Low abundance of source material
In the gut, metagenomics studies demonstrated that only ~0.1% of detectable sequences in the total microbiota were attributed to fungi (26), in contrast to the 99% that were bacterial. There are no data about what this ratio might be in the bladder or any reason to believe that this ratio would be similar. Even when considering these estimates, there is cause to doubt this conclusion, as this assessment was based on sequences identified from available annotated reference databases, in which fungi are highly underrepresented (46). Regardless, the prevailing belief is that the total abundance of microorganisms in urine is orders of magnitude less than the gut, with fungi comprising only a small percentage.
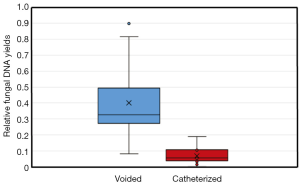
Thus, for populations of low abundance, as presumed for the urinary tract, the difficulties detailed above with appropriate sensitivity are amplified. Low numbers of fungi would result in the omission of large numbers of species from community profiling as a result of abundances below detection levels. As urine samples vary considerably from void to void even in the same individual, small volume samples may not be representative of the overall population. For example, analysis of 1 mL urine samples without controlling for the overall voided volume, urine concentration, and circumstances of collection is likely to introduce a multitude of additional biases that may influence results. We have noted significant vaginal and skin contamination in voided specimens that cannot be entirely eradicated by catheterization. Given the microbial load in the vagina, both fungal and bacterial, any vaginal contamination is likely to obscure the urinary microbiota almost completely (Figure 2). Suprapubic aspiration offers the purest examination of urine with minimal urethral, vaginal, or skin contamination (47), but due to its invasive nature, is not practical for the study of urinary fungi in larger human populations.
Bladder vs. urine
Urinary samples may not reflect the bladder microbiota. In the gut, direct comparison of mucosal biopsies to stool samples or brush biopsies demonstrate that mucosal-associated bacteria are not equally distributed between these two locations (48,49). Fungal interactions with the urothelium are entirely uncharacterized; it is possible that fungi may attach directly to the urothelium to form mixed biofilms with bacterial species or grow in soluble microcolonies in the urine, never making direct contact with the urothelium. It is even possible that fungi could remain quiescent within cells of the bladder wall, as can be seen with E. coli intracellular bacterial colonies (50). Similar mechanisms of intracellular sequestration have been observed for other fungal species in human hosts (51-54). If all of these mechanisms are in play, then the urinary microbiota would be expected to vary with urine concentration, the amount of urothelial sloughing, and intrinsic host and environmental factors, such as psychosocial stress, that alter growth conditions for these organisms.
Temporal fluctuations in urine microbial composition
As yet, no one has analyzed the stability of urinary microbial communities over time, leading to uncertainty as to whether observed differences between subjects represent stable communities with high interindividual variability, highly fluctuant intraindividual variation over time, or differences in collection methods unrelated to microbial content. Even using similar collection and processing methodologies, two studies of the oral mycobiome exhibited significant differences in the most abundant taxa (15,55), likely reflective of both the high variability between the subjects in each study as well as the different techniques utilized. Thus, a comparison of species abundance across samples using data from NGS can be misleading. With variations in collection and analytic methods, each of which introduces its own unique biases, comparison of fungal data across studies becomes challenging. All of these caveats again stress the point that these methods do not truly provide evidence of viable, stable fungal communities without confirmatory studies. While NGS studies are useful at generating an overall sense of microbial changes in disease, the limitations of these studies are many and require validation and mechanistic investigations to place the results into the context of bladder physiology.
The composition of the human mycobiome
The initial characterizations of the human mycobiome were dependent on culture-based methods, which limited these analyses to a select few species known to be pathogenic to humans, such as Candida albicans, Candida glabrata, Crytococcus neoformans, Aspergillus fumigatus, Coccidioides spp., Histoplasma spp., and Blastomyces spp. With extended culture conditions and culture-independent methods, however, we now recognize that sites thought to be sterile, such as the lung, harbor fungi (24) of astounding diversity. The composition of these communities is influenced by a multitude of factors, including age, gender, environmental variations (56), and hygiene (57). The limited fungal community profiling data available so far suggest that fungal communities may be more fluctuant than bacterial ones (58) and exhibit a greater interindividual variation than seen for bacteria (59-62). One plausible hypothesis suggests that because bacteria are numerically more abundant than fungi, bacterial communities are more stable and robust and are less influenced by environmental fluctuations.
In general, different predominant organisms are seen in different anatomic niches, but related, neighboring sites often have similar patterns (63). For example, Cladosporium, Aspergillus, and Penicillium spp. dominate in both the oral and nasal cavities (15,64), but differ from those seen in lung bronchoalveolar lavage specimens (65). The regions surrounding the urethra, however, are highly divergent in their composition; the inguinal crease skin is dominated by Malassezia spp. (31), while the vagina contains predominantly Candida spp. (66). The local environment is important as a shift in the populations can be pathogenic; a predominance of Candida in the inguinal crease is associated with dermatitis (67).
In the vagina, Candida spp. have long been recognized as colonizers of the genital epithelium without causing disease (under most circumstances). In a culture-independent analysis of the vaginal mycobiome in healthy, asymptomatic women (66), ITS1 NGS confirmed this predominance of the Ascomycota, the largest proportion of which were Candida spp. Candida was not, however, the only fungal species detected or even the predominant one; 30% of patients did not have detectable Candida at all in vagina swab specimens. As seen in other organ systems, alterations in vaginal fungal diversity were associated with pathologic conditions, such as in diabetes (68) and allergic rhinitis (69). In all of these studies, however, there are a large proportion of sequences that cannot be matched to specific taxa, which again reflects the nascent status of this field.
The urinary mycobiome
As yet, there have not been any comprehensive attempts at characterizing the urinary mycobiome reported. Low levels of Candida spp. have been detectable in urinary samples from a variety of patients, including healthy controls, using expanded quantitative urine culture (EQUC) (4-7), demonstrating their viability within urine. Using the Ibis T-5000 universal biosensor system to identify microbes, fungi were detectable in urine from patients with urological chronic pelvic pain syndromes (70). This approach recognized an increase in the number of patients with detectable fungi (3.9% vs. 15.7%) during symptomatic flares, while no significant differences in the bacterial microbiota could be identified. Of note, this detection method is only able to quantitate the Saccharomycetales, which includes Candida and Saccharomyces, but excludes many of the species implicated in other organ system pathologies. Regardless, these transformative results implicate a role for fungi in lower urinary tract symptomatology that was not previously recognized.
We have performed a preliminary, culture-independent characterization of urinary fungi from a range of asymptomatic subjects (manuscript in preparation). Utilizing NGS with amplification of the ITS1 region, we have identified a diverse population of fungi in urinary samples. As detailed in Figure 3, our results indicate that each individual exhibited a diverse fungal population without single predominant taxa. Fungi belonging to the Saccharomycetes class, which includes the Saccharomyces and Candida spp., are the only fungi documented to be detected in urine as yet (4,70). These were commonly present in our analysis but were not the most common, demonstrating the limitations of culture-based analyses at characterizing the breadth of fungal populations in urine. Similar to studies of the vagina, a proportion of the OTUs (approximately 40%) observed could not be matched to known fungal species despite comprehensive searches of multiple fungal-specific databases and the NIH GenBank sequence database of all publicly available DNA sequences.
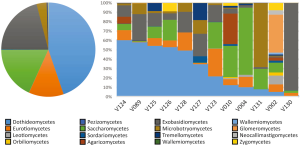
Interestingly, one patient in the analysis (#V002 in Figure 3) had a dramatically different profile of urinary fungi than the other subjects; approximately 32% of the detected OTUs were identified as belonging to the glomeromycetes, a fungal division completely absent from all other patients. In addition, the relative levels of fungal DNA found in this subject were nearly three orders of magnitude greater than the population median. After performing the analysis, it was noted that this patient was taking steroids for an unrelated skin condition at the time of sampling. While this result only details a single aberrant patient, the dramatic diversion of this subject’s resident urinary fungi from the patterns seen in the majority of other subjects suggests that the composition of the urinary mycobiome is linked to changes in systemic physiology. In this particular case, modulation of host immune status is closely tied to dramatic changes in both fungal overall quantity and community composition.
The mycobiome in health and disease
Despite the limitations in community profiling data, patterns in fungal populations begin to emerge as do their associations with organ physiology and pathology. Alterations in the mycobiome occurring in the absence of classical infections have been implicated in a wide range of diseases, such as chronic hepatitis B virus (HBV) hepatitis (25), atopic dermatitis (71), dandruff (72), IBD (73-75), cystic fibrosis (24), allergy/atopy (64), asthma (76), and psoriasis (77,78). There is no obvious pattern of fungal alterations in these diseases; pathology can be associated with either a decreased or increased diversity from that seen in the healthy state. The particular species alterations also differ in each pathology. Unlike the case of Helicobacter pylori in gastritis, the lack of simple associations of specific diseases with particular organisms underlies the complex interdependency between the mycobiome, host immune status, and other members of microbiota (including the equally neglected protozoa, viruses, and archaebacteria).
A major caveat for studies identifying associations of disease states with microbial alterations is the questionable cause logical fallacy; that is, the correlation of alterations in the microbiota with disease states does not provide evidence that these changes are causative or even impact disease progression. Changes in the mycobiome may directly lead to the generation or progression of disease, but they may also be merely a sign of altered tissue microenvironments, perhaps as a result of altered metabolism or inflammatory dysfunction. It is equally plausible that the disease state results in local environmental changes that lead to the development of an altered microbial community better adapted to the new conditions. Even if microbial changes are not involved in pathogenesis, however, specific microbial signatures may be useful as diagnostic or prognostic markers of disease.
It is unlikely that the simple introduction of a specific species or even the expansion of a class of organisms results in disease without alterations in the host that facilitate the pathology. In other diseases linked to mycobiome alterations, fungal colonization is typically coincident with an immunomodulatory state that results in dysregulated inflammation presenting without a clear etiology (79). While it is well understood that immunosuppressed patients (such as HIV+, transplant, or chemotherapy patients) are more likely to contract opportunistic fungal (and protozoal) infections than immunocompetent patients, the specific mechanisms by which immunomodulatory states lead to increased fungal pathogenesis are not well understood.
In addition to the influence of host immune status on fungal community composition, the transition to virulence of commensal fungi may also be complicated by the ability of individual fungal species to exist in both a commensal and pathogenic relationship with the host (80). The mechanisms controlling such modifications are unclear. The switch from commensal to pathogen within a single ecosystem may be associated with gene expression changes triggered by as-yet poorly understood environmental or host factors. This evidence suggests that the host response to fungi is highly complex with constant integration of complex messages from both local environments and systemic host health. Overall fungal burden, community composition, and unique fungal virulence states likely interact continually with and respond to host metabolic state and immune status as well as perturbations in other microbial communities, such that minor fluctuations in any of these factors can bias the host from symbiosis towards disease.
Future directions
Myriad challenges await in the field of urologic mycology. Most fundamental is an understanding of how the fungal mycobiota of the bladder contribute to the regulation of bladder health, function, and inflammation. While much is assumed from studies of other organ systems, nothing is known about the development/inoculation and maturation of the urinary mycobiome in early life, how the urinary mycobiota interact with neighboring communities, and the susceptibility of this ecosystem to perturbations from nutritional changes, metabolic stresses, and other host inflammatory disorders. While multiple lessons from other organ systems have demonstrated the close interplay between different kingdoms, virtually nothing is known of how urinary fungi influence urinary bacterial community composition and stability.
Hopefully, with a better understanding of the role of fungi in urinary tract disease, will come the ability to use targeted manipulation of the mycobiome for therapeutic purpose, as is becoming a reality in other organ systems. Saccharomyces boulardii is being trialed as a probiotic used in the treatment of diarrheal diseases (81,82). With recognition of the role of fungi in graft-versus-host disease, antifungals are being explored as agents to ameliorate disease development and severity (83). As we learn more about the role of fungi in urinary tract disease, novel potential therapeutic approaches, such as fungus-specific vaccinations, fungal probiotics, or targeted antifungal drugs, may become plausible adjunctive treatments.
Conclusions
While long ignored, fungi are an essential part of the human microbiota. While this field remains underdeveloped and in need of improved analytic tools and approaches, novel sequencing-based approaches to the study of fungal mycobiomes have revealed diverse communities of fungi throughout the human body, including the urinary tract. Accumulating data indicate a role for fungi in normal human physiology as well as the progression and prevention of human disease. In general, an understanding of the role of fungi in health and disease requires a systems-level, integrated approach, as opposed to a focus on specific disease-causing taxa; unlike the case of H. pylori in gastritis, microbial alterations affiliated with disease rarely are due to the presence of a single causative microbe. While fungi likely play an important role in the maintenance of microbial community structure, modulating immune function and influencing metabolism, the mechanisms by which fungi interact with other components of the microbiome and the host remain poorly characterized. These variations in fungal communities are likely closely tied to host physiology in ways we have only begun to characterize. Almost nothing is known about these interactions in the bladder; the field is ripe for future studies.
Acknowledgements
The authors would like to thank Dr. Jie Tang for his valuable insight and perspective.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 2010;10:206. [Crossref] [PubMed]
- Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science 2005;307:1915-20. [Crossref] [PubMed]
- Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe 2011;10:287-91. [Crossref] [PubMed]
- Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio 2014;5:e01283-14. [Crossref] [PubMed]
- Thomas-White KJ, Hilt EE, Fok C, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J 2016;27:723-33. [Crossref] [PubMed]
- Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014;52:871-6. [Crossref] [PubMed]
- Khasriya R, Sathiananthamoorthy S, Ismail S, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol 2013;51:2054-62. [Crossref] [PubMed]
- Siddiqui H, Nederbragt AJ, Lagesen K, et al. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol 2011;11:244. [Crossref] [PubMed]
- Fouts DE, Pieper R, Szpakowski S, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 2012;10:174. [Crossref] [PubMed]
- Groah SL, Pérez-Losada M, Caldovic L, et al. Redefining Healthy Urine: A Cross-Sectional Exploratory Metagenomic Study of People With and Without Bladder Dysfunction. J Urol 2016;196:579-87. [Crossref] [PubMed]
- Thomas-White KJ, Kliethermes S, Rickey L, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol 2017;216:55.e1-16. [Crossref] [PubMed]
- Siddiqui H, Lagesen K, Nederbragt AJ, et al. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol 2012;12:205. [Crossref] [PubMed]
- Nelson DE, Van Der Pol B, Dong Q, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One 2010;5:e14116. [Crossref] [PubMed]
- Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol 2015;12:81-90. [Crossref] [PubMed]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010;6:e1000713. [Crossref] [PubMed]
- Iliev ID, Underhill DM. Striking a balance: fungal commensalism versus pathogenesis. Curr Opin Microbiol 2013;16:366-73. [Crossref] [PubMed]
- Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med 2012;4:165rv13. [Crossref] [PubMed]
- Sanglard D. Emerging Threats in Antifungal-Resistant Fungal Pathogens. Front Med (Lausanne) 2016;3:11. [PubMed]
- Enoch DA, Ludlam HA, Brown NM. Invasive fungal infections: a review of epidemiology and management options. J Med Microbiol 2006;55:809-18. [Crossref] [PubMed]
- Brandt ME, Park BJ. Think fungus–prevention and control of fungal infections. Emerg Infect Dis 2013;19:1688-9. [Crossref] [PubMed]
- Hassall A. On the Development of Torulæ in the Urine, and on the relation of these Fungi to Albuminous and Saccharine Urine. Med Chir Trans 1853;36:23-78.9.
- Marloth RH. An apparatus for the study of matforming fungi in culture solutions. Science 1929;69:524-5. [Crossref] [PubMed]
- Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res 2012;160:258-66. [Crossref] [PubMed]
- Delhaes L, Monchy S, Fréalle E, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One 2012;7:e36313. [Crossref] [PubMed]
- Chen Y, Chen Z, Guo R, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011;70:492-8. [Crossref] [PubMed]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65. [Crossref] [PubMed]
- Gilbert JA, Hughes M. Gene expression profiling: metatranscriptomics. Methods Mol Biol 2011;733:195-205. [Crossref] [PubMed]
- Dollive S, Peterfreund GL, Sherrill-Mix S, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol 2012;13:R60. [Crossref] [PubMed]
- Yuan S, Cohen DB, Ravel J, et al. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 2012;7:e33865. [Crossref] [PubMed]
- Tang J, Iliev ID, Brown J, et al. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods 2015;421:112-21. [Crossref] [PubMed]
- Findley K, Oh J, Yang J, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013;498:367-70. [Crossref] [PubMed]
- White JR, Maddox C, White O, et al. CloVR-ITS: Automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome 2013;1:6. [Crossref] [PubMed]
- Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 2010;10:189. [Crossref] [PubMed]
- Větrovský T, Kolařík M, Žifčáková L, et al. The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Mol Ecol Resour 2016;16:388-401. [Crossref] [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31-46. [Crossref] [PubMed]
- Gilles A, Meglécz E, Pech N, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 2011;12:245. [Crossref] [PubMed]
- Bartram AK, Lynch MD, Stearns JC, et al. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol 2011;77:3846-52. [Crossref] [PubMed]
- Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621-4. [Crossref] [PubMed]
- Whiteley AS, Jenkins S, Waite I, et al. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) Platform. J Microbiol Methods 2012;91:80-8. [Crossref] [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537-41. [Crossref] [PubMed]
- Abarenkov K, Henrik Nilsson R, Larsson KH, et al. The UNITE database for molecular identification of fungi--recent updates and future perspectives. New Phytol 2010;186:281-5. [Crossref] [PubMed]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6. [Crossref] [PubMed]
- Mahé S, Duhamel M, Le Calvez T, et al. PHYMYCO-DB: a curated database for analyses of fungal diversity and evolution. PLoS One 2012;7:e43117. [Crossref] [PubMed]
- Nilsson RH, Ryberg M, Kristiansson E, et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One 2006;1:e59. [Crossref] [PubMed]
- Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014;14:405-16. [Crossref] [PubMed]
- Wolfe AJ, Toh E, Shibata N, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 2012;50:1376-83. [Crossref] [PubMed]
- Araújo-Pérez F, McCoy AN, Okechukwu C, et al. Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut Microbes 2012;3:530-5. [Crossref] [PubMed]
- Huse SM, Young VB, Morrison HG, et al. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome 2014;2:5. [Crossref] [PubMed]
- Rosen DA, Hooton TM, Stamm WE, et al. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 2007;4:e329. [Crossref] [PubMed]
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 2006;16:2161-5. [Crossref] [PubMed]
- Nicola AM, Robertson EJ, Albuquerque P, et al. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. MBio 2011;2:e00167-11. [Crossref] [PubMed]
- Charlier C, Nielsen K, Daou S, et al. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun 2009;77:120-7. [Crossref] [PubMed]
- Faro-Trindade I, Willment JA, Kerrigan AM, et al. Characterisation of innate fungal recognition in the lung. PLoS One 2012;7:e35675. [Crossref] [PubMed]
- Dupuy AK, David MS, Li L, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of malassezia as a prominent commensal. PLoS One 2014;9:e90899. [Crossref] [PubMed]
- Xu J, Mitchell TG. Geographical differences in human oral yeast flora. Clin Infect Dis 2003;36:221-4. [Crossref] [PubMed]
- Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol 2009;28:405-11. [Crossref] [PubMed]
- Dollive S, Chen YY, Grunberg S, et al. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One 2013;8:e71806. [Crossref] [PubMed]
- Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol 2008;43:831-41. [Crossref] [PubMed]
- Scupham AJ, Presley LL, Wei B, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol 2006;72:793-801. [Crossref] [PubMed]
- Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2008;2:1183-93. [Crossref] [PubMed]
- Pandey PK, Siddharth J, Verma P, et al. Molecular typing of fecal eukaryotic microbiota of human infants and their respective mothers. J Biosci 2012;37:221-6. [Crossref] [PubMed]
- Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013;5:63. [Crossref] [PubMed]
- Sellart-Altisent M, Torres-Rodríguez JM, Gómez de Ana S, et al. Nasal fungal microbiota in allergic and healthy subjects. Rev Iberoam Micol 2007;24:125-30. [Crossref] [PubMed]
- Cui L, Lucht L, Tipton L, et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 2015;191:932-42. [Crossref] [PubMed]
- Drell T, Lillsaar T, Tummeleht L, et al. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 2013;8:e54379. [Crossref] [PubMed]
- Gray M, Beeckman D, Bliss DZ, et al. Incontinence-associated dermatitis: a comprehensive review and update. J Wound Ostomy Continence Nurs 2012;39:61-74. [Crossref] [PubMed]
- Zheng NN, Guo XC, Lv W, et al. Characterization of the vaginal fungal flora in pregnant diabetic women by 18S rRNA sequencing. Eur J Clin Microbiol Infect Dis 2013;32:1031-40. [Crossref] [PubMed]
- Guo R, Zheng N, Lu H, et al. Increased diversity of fungal flora in the vagina of patients with recurrent vaginal candidiasis and allergic rhinitis. Microb Ecol 2012;64:918-27. [Crossref] [PubMed]
- Nickel JC, Stephens A, Landis JR, et al. Assessment of the Lower Urinary Tract Microbiota during Symptom Flare in Women with Urologic Chronic Pelvic Pain Syndrome: A MAPP Network Study. J Urol 2016;195:356-62. [Crossref] [PubMed]
- Zhang E, Tanaka T, Tajima M, et al. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 2011;55:625-32. [Crossref] [PubMed]
- Park HK, Ha MH, Park SG, et al. Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS One 2012;7:e32847. [Crossref] [PubMed]
- Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2016. pii: gutjnl-2015-310746.
- Hoarau G, Mukherjee PK, Gower-Rousseau C, et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn's Disease. MBio 2016;7:e01250-16. [Crossref] [PubMed]
- Chehoud C, Albenberg LG, Judge C, et al. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 2015;21:1948-56. [Crossref] [PubMed]
- Knutsen AP, Bush RK, Demain JG, et al. Fungi and allergic lower respiratory tract diseases. J Allergy Clin Immunol 2012;129:280-91. [Crossref] [PubMed]
- Paulino LC, Tseng CH, Blaser MJ. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res 2008;8:460-71. [Crossref] [PubMed]
- de Koning HD, Rodijk-Olthuis D, van Vlijmen-Willems IM, et al. A comprehensive analysis of pattern recognition receptors in normal and inflamed human epidermis: upregulation of dectin-1 in psoriasis. J Invest Dermatol 2010;130:2611-20. [Crossref] [PubMed]
- Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012;336:1314-7. [Crossref] [PubMed]
- Romani L. Immunity to fungal infections. Nat Rev Immunol 2011;11:275-88. [Crossref] [PubMed]
- Sharif MR, Kashani HH, Ardakani AT, et al. The Effect of a Yeast Probiotic on Acute Diarrhea in Children. Probiotics Antimicrob Proteins 2016;8:211-4. [Crossref] [PubMed]
- Das S, Gupta PK, Das RR. Efficacy and Safety of Saccharomyces boulardii in Acute Rotavirus Diarrhea: Double Blind Randomized Controlled Trial from a Developing Country. J Trop Pediatr 2016;62:464-70. [PubMed]
- van der Velden WJ, Netea MG, de Haan AF, et al. Role of the mycobiome in human acute graft-versus-host disease. Biol Blood Marrow Transplant 2013;19:329-32. [Crossref] [PubMed]


