Questions and challenges associated with studying the microbiome of the urinary tract
Introduction
Through direct analysis methodologies, such as microscopy, we know that there are vastly more bacterial types than that of which we can culture (1). In the clinical laboratory, bacterial propagation is mostly targeted towards specific bacterial pathogens that are associated with most causes of urinary tract infections (Escherichia coli, Staphylococcus saprophyticus, Proteus mirabilis, Enterococcus faecalis and others). However, we know little regarding the supporting roles of the non-infection-associated bacteria in these conditions, yet we know for many commensal bacteria, that their survival is dependent upon living within a bacterial community. It is estimated that only a small proportion of bacterial types from the human body can be cultured (2). However, with the more recent extended culture techniques that are being employed for urine, developed by Hilt and colleagues (3-5), a higher proportion of bacteria are likely to be cultured from the urogenital tract compared to other sites, though still likely to underestimate microbial complexity. Information mined from genomic studies will also give additional clues to the likely nutritional requirements of uncultured microorganisms that are likely to eventually assist in their propagation. Bacteria are often dependent on each other for growth and have co-evolved over millions of years in mixed communities. Culture, while widely used in the clinical environment, is laborious for microbial population analysis though still incredibly useful to study specific bacteria in much more detail, encompassing growth and metabolic characteristics, genetic regulation and allows the evaluation in models.
Next generation sequencing (NGS) methods for analysis have allowed an unparalleled amount of information to be collected. NGS for microbiota analysis typically involves the amplification of small DNA lengths (~200 bp), typically variable regions of the otherwise conserved 16S rRNA gene. When analyzed, these segments give a broad perspective of the bacterial composition, but are generally limited to a taxonomic analysis above species level. Despite this limitation, NGS has elucidated the role of the microbiome in numerous diseases and metabolic processes. Beyond microbiota analysis, “shotgun” NGS sequencing can be used for metagenomics analysis, where all the DNA from a sample is sequenced. This has the power to give perspective on the genetic capabilities that can potentially be employed by the microorganisms present. RNASeq further clarifies bacterial activity as it maps the microbial mRNA gene expression, detailing real time responses. Finally, metabolomics is a tool gaining popularity which looks at the small molecules produced by host and microbial metabolism and therefore, is useful to investigate involvement of bacteria in disease conditions. Emerging technologies will facilitate the characterization of microbiome composition with more sensitivity and ease of use. Some of these techniques, such as targeted arrays (http://www.affymetrix.com/estore/catalog/prod1060044/AFFY/Axiom+Microbiome+Arrays), will be limited in that they will only be able to determine if a specific bacterium is present or not and cannot give any information on bacteria not included on the array. However, these arrays consist of thousands of different bacterial species and will bypass the need for high level computational biology expertise to analyze and interpret, a current challenge associated with NGS.
There is no doubt that other microbiomes play a role in urology, such as those in the intestinal tract which are responsible for huge metabolic output and whose by-products not only pass locally, but also systemically through the kidneys and urinary system. This influences the urinary system health by the formation of kidney stones, reservoirs urinary tract infections (UTI), carcinogens, neuroactive substances, regulation of inflammation (transplant, chronic kidney disease), hormones (testosterone) and the urinary microbiome itself (6). The focus of this review is the more overlooked urinary microbiome. Though it has been recently characterized, its influence in the host has yet to be determined. The analysis of these bacterial communities presents unique challenges in comparison to other microbiotas, as they may be in low abundance or associated with human tissue.
What’s there and why could it be important for urology?
Microbiome of the urological system
Until relatively recently, there was no definitive proof of the existence of a microbiome above the lower urogenital tract (vagina and distal urethra) (7); however, researchers now recognize that its existence could influence urological health. We are still in the discovery phase with regard to the descriptive “snapshot” studies that are being undertaken in the urinary microbiome. Nevertheless, there is enough information to support that males and females have different microbiomes, undoubtedly influenced through ascension from the lower genital tract. Many of the studies do not entirely agree, which appears to be partially dependent upon the patient cohort (sex, disease status etc.), as reviewed by Whiteside et al. (6) There are likely to be other methodological factors that will influence this, such as sample type, storage conditions, processing and extraction techniques. In addition, the PCR primers, 16S rRNA gene variable region targeted, PCR cycle number, and sequencing platform will have a major influence, as will the bioinformatics analyses which suffer from a lack of standardization and statistically incorrect approaches.
What is the microbiome doing in the urinary system?
The drive for using newer technologies is to gain a better understanding of what microorganisms are doing at particular sites. In the lower urogenital tract, such as at the vagina, we know that certain bacteria are in fact sentinels and protectors against potentially deleterious microorganisms that may ascend and cause problems such as UTI (8,9). We do not truly understand if or how the microbiome plays a protective role in the bladder, but we assume that the lower urogenital tract is the gateway to this region. This is supported by studies demonstrating probiotic treatment reduces UTI recurrence rates (8). The numbers of commensal bacteria in the bladder are thought to be considerably less than the lower genital tract, and several still remain to be cultured (6). Even though we know bacterial numbers are lower in this region, they have the potential to outcompete pathogens for necessary nutrients, produce antimicrobial substances and stimulate the immune system. Where bacteria colonize other “exposed” human surfaces, they play a key role in the homeostasis of the mucosal environment and numerous studies detail these in skin, buccal, nasal, small intestine, bowel, lung and vaginal interactions (10-16). It therefore, makes sense to presume that even while there may be less bacteria in the urinary system than many other colonized sites, they still may play a significant role in stimulation and maintenance of the uroepithelium.
There is a complex interaction between the anatomy of the urinary tract and nervous system control for which microbes may also have a role in health and disease. During micturition, muscles are contracted and sphincters relax. To respond to the nervous and hormonal control systems, each part of the urinary tract muscles has specific receptors for the transmitters/modulators, released from nerves or generated locally. The micturition reflex is mediated by transient receptor potential cation channel nerves and others. These same neurons also convey nociceptive information (for example, bladder pain) to the central nervous system. TRPV4 (TRP subfamily V member 4) is a key player in bladder function because it is present in both the urothelial epithelium and the detrusor muscle, and it is activated by stretch and hypo-osmolar urine. It may also be activated by microorganisms in conditions such as over active bladder and even offers the potential for treatment with non-disease causing variants of pathogenic species, such as E. coli, to displace more virulent ones (17-19).
Overactive bladder (OAB) is a condition where there is a need to urinate frequently and often leads to incontinence. OAB seems to subvert the normal micturition reflex where stretch receptors in the wall of the bladder become active and send signals to spinal cord pelvic nerves. Clinicians can offer behavioral therapies (e.g., bladder training, bladder control strategies, pelvic floor muscle training, fluid management) in combination with drug management anti-muscarinics or oral β3-adrenoceptor agonists. However, few have looked at the role of bacteria in this process, though this has been investigated in urinary and stress incontinence (20-25). The mechanisms of OAB are not fully elucidated but there are potential links to microbiome involvement. These include potential contributions at the global level, where constipation is often associated with the condition and that there may be “cross talk” between gut and the bladder (26). It is interesting that UTI’s caused by different bacterial types such Enterococcus faecalis, seemingly invoke different pain responses, perhaps indicative of bacterial interference with nociceptive signaling. Indeed, E. faecalis is the second most common cause of both asymptomatic bacteriuria and symptomatic UTI, yet does not proliferate nor induce the characteristic signs of UTI (suprapubic pain, urgency, frequency and dysuria), that are associated with a “classic” E. coli UTI (27). Our preliminary investigations indicate that urogenital tract isolates, such as enterococci, are capable in vitro of producing neuroactive substances.
Interaction with the host and other microorganisms in cancer
There are many other urinary conditions that would benefit from microbiome analysis, such as the role of bacteria in the protection, induction and modulation of bladder cancer therapies. We know that commensal bacteria can easily bind several compounds that are thought to be linked to bladder cancer such as cadmium, other heavy metals and pesticides (28). If these bacteria are absent, are people at a higher risk of cancer? Also, uropathogenic E. coli can carry pathogenic islands that have been linked to colon cancer in the intestinal tract (29,30). The link between recurrent UTI and cancer is tenuous but may be a possibility in a subset of bladder cancer cases where certain inflammatory responses may invoke oncogenic responses.
Bacille Calmette-Guerin (BCG) is an attenuated strain of Mycobacterium bovis, primarily used as a vaccine against tuberculosis. However, BCG is also used intravesically to treat urothelial carcinoma lesions such as carcinoma in situ, high-grade Ta (papillary) and T1 (lamina propria invasive) lesions after transurethral resection. However, up to one third of treated patients experience recurrence or progression of the cancer. BCG is purported to work by attaching to fibronectin and integrin α5β1 then inducing an immune response (31). While there appears to be a genetic link between BCG effectiveness and Pak1-dependent micropinocytosis (32), one ponders if indigenous bladder microorganisms could be exploited to stimulate a more effective response? Our studies with fibronectin binding indicate that vaginally-derived and predominant lactobacilli such as Lactobacillus iners may be potentially superior at this compared to other species (33). Studies using non-vaginally-derived lactobacilli have been undertaken with mixed results (34-36). Another factors which may also effect this treatment includes dampening of immune responses by commensal bacterial strains, which have been effective at doing this at other mucosal sites (10).
How far up the urinary tract do bacteria go and how commonly are they found in the kidney?
Bacteria are not typically thought to be in the kidney during times of health, though it is known that some bacteria enter the blood stream and kidneys following probiotic dosing and translocation (37-39). During periods of urological imbalance, as would be encountered during urinary reflux and infection (pyelonephritis), flagellated bacteria have the ability to access the upper tract and kidneys. In more severe systemic diseases and certainly in chronic disease, there can be translocation of bacteria to the kidneys. In preliminary studies looking at the microbiome within kidney stones, it appears that bacteria can be detected both by culture and non-culture microbiological techniques (40). Interestingly, preliminary data of five pediatric subjects suggests that bacteria may be detected as far as the kidneys and may actually be present within kidney stones. The nidus for kidney stone formation and precipitation from urinary supersaturation is thought to be inflammation. The potential for bacteria to cause this is one possibility and in murine studies it was shown that the presence of calcium oxalate deposits increases pyelonephritis risk, likely due to preferential aggregation of bacteria on and around calcium oxalate crystals (40).
Technical challenges
Correct approaches for sampling the urogenital microbiome
Microbiome sampling from fecal and tissue material is complicated by effective separation of bacteria from the sample matrix and removal of PCR inhibitors. Contrary to this, urine is quite amenable to microbiome analysis, as it is easy to collect, contains very little matrix material and fewer PCR inhibitors. Although bacteria are low in numbers, they can be concentrated easily by centrifuging a larger sample volume. Mid-stream urine samples are thought to be representative of the bladder in both males and females, whereas first void is more typical of the distal urethra. Allowances still need to be made for urine samples contaminated with trace amounts from the lower genital tract microbiota. Urine typically contains high concentrations of salt, which allows the preservation of bacteria and nucleic acids for some time, though from our laboratory’s perspective, bacteria should be pelleted and separated from the liquid phase prior to frozen storage. Mid-stream urine can also be used as a suitable material to study microbiome interactions in the bladder such as urinary tract infection, interstitial cystitis, bladder pain syndrome and bladder cancer, amongst others, while transurethral catheterisation and suprapubic aspiration remains as more invasive procedures but are less vulnerable to genital contamination.
Clinical contamination of samples
Other tissues and materials (kidney stones, stents, catheters, and mesh) are often passed through the urogenital tract, this may contaminate samples with bacteria. Surgical opportunities to recover material via non-urogenital entry are less common (i.e., percutaneous nephrolithotomy for more clinically difficult kidney stones) but may provide a better avenue for more accurate microbiome analyses. These patients typically undergo a flank incision, or for renal cancer and nephrectomy procedures may require an open surgical method. Surgeries that enter the urinary system above the lower urogenital tract are likely to avoid such contamination, although in laparoscopic surgery the skin microbiome can be a risk-factor for bacterial contamination of samples (Figure 1). On the other hand, surgical instruments are all sterile but not DNA-free so a background DNA noise is almost inevitable and an environment control sample is important. It is also particularly useful to discuss with the surgeon the exact nature of the material and strategies for its removal to prevent contamination. Clearly in surgeries for oncology purposes tissue is often well documented (tumor or adjacent tissue, position, size etc.) but not in others such as stone removal which might involve whole stones or only fragments. If researchers are considering studying bacterial composition in relation to stone disease, the initial location of clinical material that is removed is an important factor to consider. Operating rooms may be clean, but they are not totally sterile. Appropriate control samples should be collected from these environments to allow these potential contaminants to be deducted from the downstream analyses.
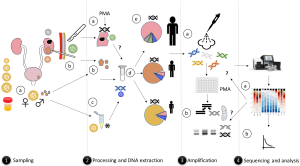
Surgical intervention is not the only route to collect samples for microbiome analyses. Extracorporeal shock wave lithotripsy (ESWL) is a noninvasive procedure which allows small stone fragments to be retrieved in urine and the post lithotripsy urine could be potentially indicative of the microbiome from this site. However, distinguishing them from the lower urinary tract bacterial DNA can be difficult. Procedures, such as ESWL, may also disrupt biofilms at lower sites (i.e., bladder) resulting in the release of non-targeted microorganisms. When investigating these samples for bacteria, thorough knowledge of where the sample of tissue or stone was obtained is required. Kidney stones, especially larger ones, are expelled in pieces, potentially increasing the risk of contamination. In addition, while we theorize that bacteria may be the nidus for stones or other conditions in tissue such as cancer, the samples may not necessarily represent the bacterial compositions at these exact sites.
DNA extraction and amplification bias
While the use of 16S rRNA gene sequencing has revolutionized our understanding of the microbiome at a number of anatomical sites, it has a number of limitations regarding the inability to differentiate between DNA originating from either live or dead cells. It is also highly vulnerable to contamination during sample collection and within laboratory processing. Next generation sequencing of the microbiome provides information about relative abundance of each component in a sample, but cannot estimate the absolute number of DNA copies. Nonspecific amplification by V4/V5 16S rRNA PCR primers in the presence of high quantities of mammalian DNA is known to occur and can incorrectly overshadow the 16S rRNA gene contribution of the microbiome.
In samples with low microbial abundance, care must be taken to prevent cross amplification of the host’s mitochondrial DNA which has some sequence similarities to the 16S rRNA bacterial gene. This is a particular concern when there are few bacteria and the “background” amplification is forced into the foreground with regards to analysis. This problem is further compounded when the number of PCR cycles is increased. These are problems rarely encountered in samples that have high abundance because it is typically a minor component of the sequencing reads. When isolating bacterial DNA from tissue samples (mixed human and microbial DNA), the human DNA competes with the bacterial DNA during the isolation procedure, decreasing the amount of bacterial DNA recovered. Many DNA extraction kits rely upon silica gel membranes to bind free DNA from the sample. This includes the MoBIO PowerSoil® kit which is rapidly becoming the standard kit for the human and earth microbiome projects. Such columns have maximum binding capacity of 20 µg of DNA. Given the average bacterial genome size (2–6 Mb) and the human genome (~3,000 Mb), a silica column will reach the limit of isolation after ~109 copies of bacterial gDNA but will only need to bind ~106 copies of human DNA. In actuality, this is a small quantity given 1g of tissue equates to approximately 108–109 copies of human DNA, thus DNA from a small amount of mammalian tissue will rapidly bring a DNA purification column to capacity, not allowing any bacterial DNA to be isolated despite being present.
Tools to mitigate bias and contamination
Treatment of samples by propidium monoazide (PMA), a photoactivated DNA binding agent which can reduce surface contamination of tissues, other samples and reagents. This technique has yet to be widely employed in the processing of samples for microbiome studies but offers great promise in reducing DNA contamination. PMA binds both free DNA and DNA contained within unmaintained membranes (dead bacteria), arresting PCR amplification of the PMA-bound DNA. Washing samples with sterile and nucleic acid free water or ethanol will also reduce the potential of environmental surface contamination. Combined with other tools such as quantitative PCR, the estimation of DNA copies representing bacterial cell numbers can be determined.
Host DNA depletion steps can be performed by specific kits to maximize bacterial DNA yield, though these have not been widely used to date. Some of these kits facilitate enrichment of microbial DNA from samples by selective binding and removing CpG-methylated (human) DNA. According to manufacturer’s information, microbial diversity remains intact after enrichment. (https://www.neb.com/products/e2612-nebnext-microbiome-dna-enrichment-kit).
Conclusions
There is still much to learn about the microbiome’s role in urology and its functional role in health and disease. While there are many useful “snapshot” studies of the urinary microbiota, few have been conducted in a longitudinal manner. These types of studies are required to understand the urinary microbiome’s development and its influence on health and disease from birth to death. Early changes in the microbiome may have consequences later in life. Bacteria are found ubiquitously, even in sites once considered sterile, including the urinary system. In low abundance samples, including the urinary tract, careful mitigation of numerous contaminating factors is required including the dominance of host DNA in some samples. However, when there is a greater understanding of the role the microbiome plays in this region, we will be able to modulate its response with the intent of improving health outcomes.
Acknowledgements
The developmental work described in this review was largely supported by W. Garfield Weston Foundation, Toronto, Canada.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Moter A, Göbel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods 2000;41:85-112. [Crossref] [PubMed]
- Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 2007;9:2125-36. [Crossref] [PubMed]
- Price TK, Dune T, Hilt EE, et al. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol 2016;54:1216-22. [Crossref] [PubMed]
- Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014;52:871-6. [Crossref] [PubMed]
- Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio 2014;5:e01283-14. [Crossref] [PubMed]
- Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol 2015;12:81-90. [Crossref] [PubMed]
- Thomas-White K, Brady M, Wolfe AJ, et al. The bladder is not sterile: History and current discoveries on the urinary microbiome. Curr Bladder Dysfunct Rep 2016;11:18-24. [Crossref] [PubMed]
- Beerepoot M, Geerlings S. Non-Antibiotic Prophylaxis for Urinary Tract Infections. Pathogens 2016;5:E36. [Crossref] [PubMed]
- Beerepoot M, Ter Riet G, Geerlings SE. Lactobacilli vs Antibiotics to Prevent Recurrent Urinary Tract Infections: An Inconclusive, Not Inferior, Outcome-Reply. Arch Intern Med 2012;172:1690-4. [Crossref] [PubMed]
- Cosseau C, Devine DA, Dullaghan E, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun 2008;76:4163-75. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol 2015;309:L1047-55. [PubMed]
- SanMiguel A, Grice EA. Interactions between host factors and the skin microbiome. Cell Mol Life Sci 2015;72:1499-515. [Crossref] [PubMed]
- Lopetuso LR, Scaldaferri F, Franceschi F, et al. The gastrointestinal microbiome - functional interference between stomach and intestine. Best Pract Res Clin Gastroenterol 2014;28:995-1002. [Crossref] [PubMed]
- Pfefferle PI, Renz H. The mucosal microbiome in shaping health and disease. F1000Prime Rep 2014;6:11. [Crossref] [PubMed]
- Lee JT, Frank DN, Ramakrishnan V. Microbiome of the paranasal sinuses: Update and literature review. Am J Rhinol Allergy 2016;30:3-16. [Crossref] [PubMed]
- Hummelen R, Macklaim JM, Bisanz JE, et al. Vaginal microbiome and epithelial gene array in post-menopausal women with moderate to severe dryness. PLoS One 2011;6:e26602. [Crossref] [PubMed]
- Encyclopedic Reference of Molecular Pharmacology. Springer Berlin Heidelberg 2004:931.
- Rudick CN, Taylor AK, Yaggie RE, et al. Asymptomatic bacteriuria Escherichia coli are live biotherapeutics for UTI. PLoS One 2014;9:e109321. [Crossref] [PubMed]
- Rosen JM, Klumpp DJ. Mechanisms of pain from urinary tract infection. Int J Urol 2014;21 Suppl 1:26-32. [Crossref] [PubMed]
- Thomas-White KJ, Kliethermes S, Rickey L, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol 2017;216:55.e1-55.e16. [Crossref] [PubMed]
- Karstens L, Asquith M, Davin S, et al. Does the Urinary Microbiome Play a Role in Urgency Urinary Incontinence and Its Severity? Front Cell Infect Microbiol 2016;6:78. [Crossref] [PubMed]
- Cox L, Rovner ES. Lower urinary tract symptoms in women: epidemiology, diagnosis, and management. Curr Opin Urol 2016;26:328-33. [Crossref] [PubMed]
- Thomas-White KJ, Hilt EE, Fok C, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J 2016;27:723-33. [Crossref] [PubMed]
- Pearce MM, Zilliox MJ, Rosenfeld AB, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol 2015;213:347.e1-11. [Crossref] [PubMed]
- Fenner A. Incontinence: A role for the microbiome in urge incontinence? Nat Rev Urol 2014;11:486. [Crossref] [PubMed]
- Braundmeier-Fleming A, Russell NT, Yang W, et al. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci Rep 2016;6:26083. [Crossref] [PubMed]
- Horsley H, Malone-Lee J, Holland D, et al. Enterococcus faecalis subverts and invades the host urothelium in patients with chronic urinary tract infection. PLoS One 2013;8:e83637. [Crossref] [PubMed]
- Monachese M, Burton JP, Reid G. Bioremediation and tolerance of humans to heavy metals through microbial processes: a potential role for probiotics? Appl Environ Microbiol 2012;78:6397-404. [Crossref] [PubMed]
- Nougayrède JP, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006;313:848-51. [Crossref] [PubMed]
- Bronowski C, Smith SL, Yokota K, et al. A subset of mucosa-associated Escherichia coli isolates from patients with colon cancer, but not Crohn's disease, share pathogenicity islands with urinary pathogenic E. coli. Microbiology 2008;154:571-83. [Crossref] [PubMed]
- Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol 2014;11:153-62. [Crossref] [PubMed]
- Redelman-Sidi G, Iyer G, Solit DB, et al. Oncogenic activation of Pak1-dependent pathway of macropinocytosis determines BCG entry into bladder cancer cells. Cancer Res 2013;73:1156-67. [Crossref] [PubMed]
- McMillan A, Macklaim JM, Burton JP, et al. Adhesion of Lactobacillus iners AB-1 to human fibronectin: a key mediator for persistence in the vagina? Reprod Sci 2013;20:791-6. [Crossref] [PubMed]
- Seow SW, Rahmat JN, Mohamed AA, et al. Lactobacillus species is more cytotoxic to human bladder cancer cells than Mycobacterium Bovis (bacillus Calmette-Guerin). J Urol 2002;168:2236-9. [Crossref] [PubMed]
- O'Donnell MA. Does the probiotic L. casei help prevent recurrence after transurethral resection for superficial bladder cancer? Nat Clin Pract Urol 2008;5:526-7. [Crossref] [PubMed]
- Naito S. Comment on "Does the probiotic L. casei help prevent recurrence after transurethral resection for superficial bladder cancer?". Nat Clin Pract Urol 2009;6:E5. [Crossref] [PubMed]
- Anders HJ, Andersen K, Stecher B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int 2013;83:1010-6. [Crossref] [PubMed]
- Pahl MV, Vaziri ND. The Chronic Kidney Disease - Colonic Axis. Semin Dial 2015;28:459-63. [Crossref] [PubMed]
- Sabatino A, Regolisti G, Brusasco I, et al. Alterations of intestinal barrier and microbiota in chronic kidney disease. Nephrol Dial Transplant 2015;30:924-33. [Crossref] [PubMed]
- Barr-Beare E, Saxena V, Hilt EE, et al. The Interaction between Enterobacteriaceae and Calcium Oxalate Deposits. PLoS One 2015;10:e0139575. [Crossref] [PubMed]


