RNAs that make a heart beat
Action potentials drive heart beat
The beating of a heart is a symbol of life and vitality. The ability of the heart to pump blood throughout the body is essential for bringing oxygen and nutrients to peripheral tissues and removing carbon dioxide and other wastes. Heart beats power this flow of blood. Each time the heart beats, arteries expand and fill with blood; the pauses between the beats allow for constriction followed by a re-expansion with the next heart beat.
The ability of the heart to beat relies on the conversion of electrical to mechanical energy (1). The electrical basis for heart beats are action potentials that start in the specialized pacemaker cells of the heart, and are then transmitted to the atrial and ventricular heart muscle cells, cardiac myocytes, via the passage of ions between cells through gap junctions. Action potentials involve the flow of ions into and out of cardiac myocytes through voltage-gated channels. These channels open and close depending on the membrane potential, the difference in electrical potential on the inside of the cell compared with the outside (Figure 1). During an action potential, the opening and closing of one set of channels leads to a change in membrane potential, thereby allowing a different set of channels to open. These changes lead to entrance or exit of different ions, and subsequent changes in transmembrane potential (3-7).
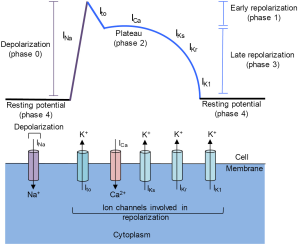
During cardiac action potentials, there is a flux of sodium (Na+) into the cell, along with an inward flux of calcium (Ca+), that leads to the depolarization, followed by outward potassium (K+) currents that repolarize the cell (1,8-11). An action potential is divided into several phases (1,9-12). In phase 4, the resting phase, the membrane potential of a ventricular cardiomyocyte is about −90 mV because, like most cells, ventricular cardiomyocytes have a more negative charge inside than outside of the cell. The membrane potential is established largely by potassium channels that allow for a loss of positively charged potassium ions from the inside of the cell resulting in a negative membrane potential. In addition, sodium-potassium pumps actively transport three sodium ions out of the cell and two potassium ions into the cell, both against a concentration and electrical gradient, thus maintaining the concentrations of both ions and preserving the voltage polarization.
Phase 0 is the depolarization phase that initiates the action potential. An action potential can result from an electrical stimulation, or, in the case of the pacemaker cells of the heart, from spontaneous automaticity of the pacemaker cells due to a slowly depolarizing Na channel that generates a pacemaker current called the funny current (10). In ventricular myocytes, rapidly activating, phase 0, voltage-gated channels specific for sodium ions open. Sodium ions are more abundant outside than inside the cell, and opening these channels results in a rush of positively charged sodium ions into the cell (INa). The flooding of sodium ions changes the transmembrane potential from negative to positive, resulting in depolarization of the membrane to about +20 mV.
The subsequent phases involve a repolarization of the membrane as positive ions flow outward from the cell. In the next phase, phase 1, the sodium channels close and voltage-gated potassium (Kv) channels open, creating an outward potassium current Ito. The rapid efflux of positively charged potassium ions results in a sharp decrease in the membrane potential from ~+20 mV to ~+10 mV at the end of phase 1. Phase 1 of an action potential can influence the height and duration of the ensuing phases (12,13).
The membrane repolarization of phase 1 activates voltage-gated calcium currents (ICa). In phase 2, a plateau phase, there is a balance between a slow inward flux of calcium ions and a reduced outward flux of potassium ions through a different set of potassium channels, the slow delayed rectifier potassium channels (IKs). The membrane potential in phase 2 ends at ~−20 mV. The calcium ions that enter the cell in phase 2 act as triggers for contraction of the myofilaments of the heart muscle.
In phase 3, the calcium channels close. The rapid delayed rectifier potassium current (IKr) contributes to the rapid potassium efflux that creates phase 3 of repolarization. Finally, an inward delayed rectifier potassium current (IK1) helps establish and maintain the cell transmembrane potential (phase 4). The IK1 current represents the current that must be overcome to establish the next action potential.
A cardiac action potential lasts ~200 msec. After an action potential, there is a refractory period of ~250 msec in which another action potential is unable or less likely to occur. After the refractory period, the cell returns to the resting phase and can respond to signals that evoke a subsequent action potential.
Voltage-gated potassium channels are tightly regulated to control myocyte function and arrhythmia
Voltage-gated channels allow fast and selective ion permeation that is regulated by opening and closing of a pore through a mechanism that senses transmembrane voltage (14). Voltage-gated channels are composed of alpha pore-forming subunits and accessory subunits (1,15). The pore-forming alpha subunits contain multiple transmembrane domains. The channels can be homotetrameric or heterotetrameric with four voltage-sensitive alpha subunits arranged surrounding a common center that, when open, serves as a pore for ions (15).
The Kv channel has six transmembrane domains, with both amino and carboxy termini localized within the cell (14). The two transmembrane domains at the carboxy terminus of the protein form the pore for potassium ion flux. The transmembrane domain adjacent to the pore pore-forming domains contains positively charged arginine and lysine residues and moves in response to membrane depolarization, thereby mediating the voltage-dependent opening of the channel (15).
Through extensive gene duplication events, the human genome encodes 40 voltage-gated potassium channels (12). The expression and regulation of these potassium channels, for instance, in different positions within the heart, can lead to changes in action potential amplitude, duration, waveforms, and rhythmicity (8,12). The transient voltage-gated potassium current (Ito) in phase one is sometimes divided into Ito,fast and Ito,slow based on the rate of recovery (20–100 ms for the fast current and seconds for the slow current) (11). The pore-forming subunits of Ito,fast are Kv4.2 and Kv4.3, which are encoded by the KCND2 and KCND3 genes. These proteins can likely form pores as homomers or heteromers (16,17).
Heart beats that deviate from the normal rhythm are called arrhythmias (11). Heart failure, a common cause of death worldwide, is frequently associated with arrhythmias and electrical instability. Abnormalities in the repolarization phase of action potentials due to heart failure can contribute to arrhythmias (11,18). Dramatic changes in the levels and properties of myocardial potassium currents have been observed with cardiac disease (8), including myocardial infarction, the death and destruction of heart muscle as a result of lack of oxygen. In canine models of myocardial infarction, potassium currents are down-regulated in cells in the infarcted zone (19-21), that is, the portion of tissue that is dying or dead due to lack of blood supply. The down-regulation is most pronounced within days following the infarct and returns to normal over the course of two months. These findings highlight the physiological importance of the regulation of potassium channels in the heart.
miRNAs regulate cardiac function
The regulation of potassium channels in normal physiology or heart disease can occur through multiple mechanisms. Transcriptional regulation plays an important role in the proper expression of different potassium channel components (8). Other mechanisms that regulate potassium channel activity include splicing, RNA editing, and post-translational modifications such as phosphorylation (8,11). In addition, microRNAs have been demonstrated as post-transcriptional regulators of potassium channel expression (2).
Mature microRNAs are small 22–26 nucleotide single-stranded endogenously encoded RNAs (22,23). Originally transcribed as pri-miRNAs, they are processed by the RNAse III Drosha, Dgcr8, and other factors to form a hairpin of ~70 nucleotides in the nucleus. The hairpins are then processed in the cytoplasm by Dicer to give rise to mature miRNAs (24-26). These miRNAs can associate with Argonaute proteins in complexes called RNA-induced silencing complexes (RISC). miRNAs in the RISC complex can anneal to mRNA transcripts with similar base pair sequences in their 3’UTRs or coding regions (27,28). In most cases, miRNA targeting leads to the degradation of transcripts or inhibition of their translation (29,30).
Families of miRNAs including miR-1, miR-29, miR-15 and miR-208 have been demonstrated to respond to cardiac stress and play a role in controlling heart function (2,8,11). Van Rooij and colleagues found that microRNAs are up- and down-regulated in cardiac tissue from mice undergoing cardiac stress (31), and in response to myocardial infarction (32). Downregulation of miR-29, a microRNA that targets extracellular matrix proteins (33-35), was found to contribute to the fibrotic response post myocardial infarction (32). Ikeda and colleagues extended these studies to humans, and reported reproducible changes in miRNA levels in human patients with heart failure (36). The functional importance of miRNAs for maintaining healthy hearts has been demonstrated by cardiomyocyte-specific deletion of either Dicer or Dgcr8. Deletion of Dicer in 3-week-old mouse cardiomyocytes resulted in arrhythmias and lethality, while deletion of Dicer in the hearts of adult mice led to severe heart failure (37). Similarly, perinatal deletion of Dgcr8 in mice resulted in severe and lethal heart failure (38).
Overexpression and loss-of function studies have revealed roles for individual miRNAs in heart disease (31,39,40). For example, miR-208 is expressed from the intron of myosin heavy chain 6 and is expressed specifically in cardiac and slow skeletal muscle (41). miR-208-knockout mice respond to cardiac stress with reduced fibrosis and hypertrophy (heart cell enlargement) (42). Inhibition of miR-208a in rats fed a high salt diet resulted in improved survival and reduced cardiac fibrosis (43). As another example, miR-15 is induced in the infarcted region of the heart in response to ischemia-reperfusion injury in mice and pigs (39). Systemic delivery of inhibitors of miR-15 family members in mice with myocardial infarctions reduced the size of the infarcted region and enhanced cardiac function (39).
In addition to miRNAs that affect the survival of myocardial cells, there are also examples in which miRNAs specifically target mRNAs encoding ion channels. miR-1 is expressed in heart and skeletal muscle and is overexpressed in individuals with coronary artery disease (44). Homozygous deletion of one of two miR-1 genes (miR-1-2) in mice resulted in mortality of 50% of the offspring by developmental abnormalities (45). Most of the survivors died from sudden cardiac death caused by arrhythmias (45). Several targets of miR-1 could explain these findings. In addition to the notch ligand delta (46) and the Rho GTPase Cdc42 (47), miR-1 also regulates Iroquois related homeobox 5 (Irx5), a transcriptional repressor of the Ito component potassium voltage-gated channel, Shal-related family, member 2 (Kcnd2) (45). miR-1 also post-transcriptionally represses potassium voltage-gated channel subfamily J member 2 (Kcnj2), which encodes the potassium channel subunit Kir2.1, a component of the inwardly rectifying potassium channel (44), and gap junction protein alpha 1 (Gja), which encodes connexin 43, a connexin important for cardiac gap junctions (44). Consistent with an important role for miR-1 in regulating action potentials, introduction of miR-1 was found to exacerbate arrhythmias, while introduction of miR-1 antisense inhibitors in rat hearts undergoing myocardial infarction reduced arrhythmias (44).
miR-223-3p is induced with myocardial infarction, targets Kv4.2 in its coding region, and promotes arrhythmia
In a recent issue of Annals of Translational Medicine: Cellular Physiology and Biochemistry, Liu and colleagues now report that miR-223-3p regulates cardiac function (48). Inspired by previous studies showing that miR-223-3p is upregulated in hearts after myocardial infarction (32), Liu and colleagues induced myocardial infarction in rats by tying off the artery that supplies the heart with blood (a method called left anterior descending artery ligation). This procedure resulted in arrhythmias in the rats and mortality in 40% of animals. Heart tissue surrounding the infarct area contained strongly elevated levels of miR-223-3p and miR-1.
Liu and co-authors recognized that miR-223-3p is complementary to, and therefore has the potential to bind to, the coding sequence of the Kcnd2 transcript. Kcnd2 encodes Kv4.2, the alpha subunit of the voltage-gated transient outward potassium channel that carries Ito in the rat during phase 1 of an action potential (48) (Figure 2). Consistent with this hypothesis, Liu and colleagues found that Kv4.2 protein levels were lower in the area around the rat’s infarcted heart tissue. Further, down-regulation of Kv4.2 was found to be associated with reduced Ito flux. To determine whether Kv4.2 is a bonifide target of the miR-223-3p miRNA, the authors subcloned the coding sequence of Kv4.2 into a luciferase-expressing plasmid to generate a luciferase-Kcnd2 chimeric vector, and transfected the vector into neonatal rat ventricular cardiomyocytes. Co-transfection of miR-223-3p, but not a negative control, substantially suppressed luciferase activity. Finally, Liu and colleagues transfected an inhibitor of miR-223-3p into the ventricular cardiomyocytes of rats and found that the inhibitor significantly reduced the incidence of arrhythmias after acute myocardial infarction. The findings, taken together, support an important role for miR-233-3p as a regulator of cardiac action potentials after a myocardial infarction in rats.
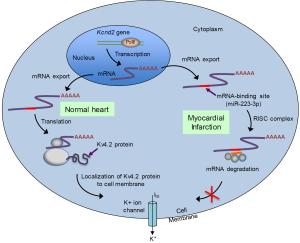
miR-223-3p inhibition as a therapy for myocardial infarction-induced arrhythmia
The findings suggest that inhibiting the induction of miR-223-3p during a myocardial infarction could normalize action potentials and benefit patients. Indeed, potassium channels are being recognized as important therapeutic targets and strategies to inhibit potassium channels are being explored as treatment for arrhythmias and other conditions including seizures, pain, and Alzheimer’s disease (49). Among the strategies to inhibit potassium channels, miRNA inhibitors are particularly appealing because they recognize specific mRNA sequence (50). Indeed, preclinical models are being developed by miRagen/Servier and other companies for the inhibition of miRNAs associated with cardiovascular disease (50,51).
The possibility of a new therapeutic strategy for myocardial infarction raises several issues. First, upregulation of miR-223-3p in human hearts undergoing myocardial infarction and miR-223-3p targeting of human KCND2 would need to be established.
Next, miR-223, the same miRNA discovered to be associated with arrhythmia, has also been associated with cardioprotection. In one study, both arms of miR-223, 3p and 5p, were discovered to be induced after ischemia reperfusion in mouse hearts (52). But in this study, as opposed to Liu et al., overexpressing a precursor miRNA containing both strands (5’ and 3’) of miR-223 was associated with better contractility and reduced necrosis after myocardial ischemia (52). Transgenic mice with a knockout of the miR-223 locus exhibited aggravated ischemia-reperfusion-induced cardiac dysfunction and more cell death (52). This study identified two cell death receptors, Tnfr1 and Dr6, as miR-223 targets in mouse hearts (52). In another study, downregulation of miR-223 occurred in the hearts of mice with severe sepsis (53). In this study, sepsis-induced mortality, inflammation and cardiac dysfunction were exacerbated in mice with knockout of miR-223-3p. Semaphorin 3A was identified as a candidate gene mediating this effect. These studies raise concerns that inhibition of miR-223-3p in hearts would have negative consequences.
Another issue is that Liu and colleagues focus on Kcnd2, which is well-established as a mediator of outward potassium current in rodent heart left ventricle apex cells. However, heart rates and action potential duration differ between species. While the phases of a heart beat are similar in all mammals, human hearts beat ~60 times per minute while rodent hearts beat ~600 times each minute (11). Until recently, the prevailing paradigm was that larger mammals such as dogs and humans rely on KCND3-encoded channels, while rodents use both Kcnd2 and Kcnd3-encoded channels and thereby achieve more rapid depolarization required for their fast heart beats (11,54,55). However, a recent discovery of a gain-of-function point mutation in KCND2 in a human patient with sudden cardiac arrest suggests KCND2-encoded proteins may be important for proper action potentials in humans as well (54). Thus, additional studies will be needed to determine the importance of Kv4.2 in human action potentials.
In addition, achieving the correct level of Kv4.2 may be challenging. Adding inhibitors of miR-223-3p would be expected to increase Kv4.2 and thus Ito. Established anti-arrhythmia drugs inhibit Ito (56), supporting the importance of Ito in establishing proper action potentials. However, inhibition of miR-223-3p could result in excess Kv4.2, elevated Ito, and arrhythmias, raising concern that miRNAs targeting Kv4.2 may increase rather than reduce arrhthyimia.
Other concerns relate to the characteristics of miRNAs. miRNAs tend to have modest effects on the target gene, off-target effects, and more pleiotropic effects than expected. For example, an anti-miR against the cardiac-specific miR-208a was unexpectedly discovered to prevent systemic phenotypes of obesity and metabolic syndrome (57). In addition, because miRNA and miRNA inhibitors are not expected to affect proteins that exist, but rather affect the levels and translation of mRNA transcripts that code for new protein, determining the pharmacokinetics of any proposed therapy will be important. Finally, developing anti-miRNAs that are not rapidly degraded and will accumulate to therapeutic levels in the heart will also be necessary (43).
In summary, proper regulation of the ion channels that control action potentials is critical for normal heart beat. Dysregulation of these channels can contribute to arrhythmia-induced mortality. miRNAs represent an emerging mechanism for regulating the expression levels of ion channel components, and are being developed as novel emerging therapeutic targets for heart disease.
Acknowledgements
The authors would like to thank Kalyanam Shivkumar and Marmar Vaseghi for helpful comments.
Funding: This work was funded by grants to HAC National Institute of General Medical Sciences R01 GM081686 and National Institute of General Medical Sciences R01 GM0866465. HAC is a member of the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research, the Jonsson Comprehensive Cancer Center, the UCLA Molecular Biology Institute, the UCLA Institute for Quantitative and Computational Biology, and the UCLA Bioinformatics Interdepartmental Program.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Grant AO. Cardiac ion channels. Circ Arrhythm Electrophysiol 2009;2:185-94. [Crossref] [PubMed]
- Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl Res 2013;161:381-92. [Crossref] [PubMed]
- Hodgkin AL, Huxley AF, Katz B. Measurements of current-voltage relations in the membrane of the giant axon of Loligo. J Physiol (Lond) 1952;116:424-48. [Crossref] [PubMed]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol (Lond) 1952;117:500-44. [Crossref] [PubMed]
- Woodbury LA, Woddbury JW, Hecht HH. Membrane resting and action potentials from single cardiac muscle fibers. Circulation 1950;1:264-6. [Crossref] [PubMed]
- Weidmann S. Effect of current flow on the membrane potential of cardiac muscle. J Physiol (Lond) 1951;115:227-36. [Crossref] [PubMed]
- Hutter OF, Trautwein W. Vagal and sympathetic effects on the pacemaker fibers in the sinus venosus of the heart. J Gen Physiol 1956;39:715-33. [Crossref] [PubMed]
- Yang KC, Nerbonne JM. Mechanisms contributing to myocardial potassium channel diversity, regulation and remodeling. Trends Cardiovasc Med 2016;26:209-18. [Crossref] [PubMed]
- Qu Z, Hu G, Garfinkel A, et al. Nonlinear and Stochastic Dynamics in the Heart. Phys Rep 2014;543:61-162. [Crossref] [PubMed]
- Bartos DC, Grandi E, Ripplinger CM. Ion Channels in the Heart. Compr Physiol 2015;5:1423-64. [Crossref] [PubMed]
- Schmitt N, Grunnet M, Olesen SP. Cardiac potassium channel subtypes: new roles in repolarization and arrhythmia. Physiol Rev 2014;94:609-53. [Crossref] [PubMed]
- Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev 2005;85:1205-53. [Crossref] [PubMed]
- Delmar M. Role of potassium currents on cell excitability in cardiac ventricular myocytes. J Cardiovasc Electrophysiol 1992;3:474-86. [Crossref]
- Yellen G. The voltage-gated potassium channels and their relatives. Nature 2002;419:35-42. [Crossref] [PubMed]
- Birnbaum SG, Varga AW, Yuan LL, et al. Structure and function of Kv4-family transient potassium channels. Physiol Rev 2004;84:803-33. [Crossref] [PubMed]
- Liu J, Kim KH, Morales MJ, et al. Kv4.3-Encoded Fast Transient Outward Current Is Presented in Kv4.2 Knockout Mouse Cardiomyocytes. PLoS One 2015;10:e0133274. [Crossref] [PubMed]
- Guo W, Jung WE, Marionneau C, et al. Targeted deletion of Kv4.2 eliminates I(to,f) and results in electrical and molecular remodeling, with no evidence of ventricular hypertrophy or myocardial dysfunction. Circ Res 2005;97:1342-50. [Crossref] [PubMed]
- Näbauer M, Kääb S. Potassium channel down-regulation in heart failure. Cardiovasc Res 1998;37:324-34. [Crossref] [PubMed]
- Dun W, Baba S, Yagi T, et al. Dynamic remodeling of K+ and Ca2+ currents in cells that survived in the epicardial border zone of canine healed infarcted heart. Am J Physiol Heart Circ Physiol 2004;287:H1046-54. [Crossref] [PubMed]
- Jiang M, Cabo C, Yao J, et al. Delayed rectifier K currents have reduced amplitudes and altered kinetics in myocytes from infarcted canine ventricle. Cardiovasc Res 2000;48:34-43. [Crossref] [PubMed]
- Pinto JM, Boyden PA. Reduced inward rectifying and increased E-4031-sensitive K+ current density in arrhythmogenic subendocardial purkinje myocytes from the infarcted heart. J Cardiovasc Electrophysiol 1998;9:299-311. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11:441-50. [Crossref] [PubMed]
- Zamore PD, Tuschl T, Sharp PA, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000;101:25-33. [Crossref] [PubMed]
- Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001;106:23-34. [Crossref] [PubMed]
- Hutvágner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001;293:834-8. [Crossref] [PubMed]
- Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91-105. [Crossref] [PubMed]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A 2008;105:14879-84. [Crossref] [PubMed]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008;9:102-14. [Crossref] [PubMed]
- Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466:835-40. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255-60. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. [Crossref] [PubMed]
- Suh EJ, Remillard MY, Legesse-Miller A, et al. A microRNA network regulates proliferative timing and extracellular matrix synthesis during cellular quiescence in fibroblasts. Genome Biol 2012;13:R121. [Crossref] [PubMed]
- Cushing L, Kuang P, Lu J. The role of miR-29 in pulmonary fibrosis. Biochem Cell Biol 2015;93:109-18. [Crossref] [PubMed]
- Maurer B, Stanczyk J, Jungel A, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 2010;62:1733-43. [Crossref] [PubMed]
- Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics 2007;31:367-73. [Crossref] [PubMed]
- da Costa Martins PA, Bourajjaj M, Gladka M, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 2008;118:1567-76. [Crossref] [PubMed]
- Rao PK, Toyama Y, Chiang HR, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 2009;105:585-94. [Crossref] [PubMed]
- Hullinger TG, Montgomery RL, Seto AG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 2012;110:71-81. [Crossref] [PubMed]
- Mutharasan RK, Nagpal V, Ichikawa Y, et al. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol 2011;301:H1519-30. [Crossref] [PubMed]
- van Rooij E, Quiat D, Johnson BA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 2009;17:662-73. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007;316:575-9. [Crossref] [PubMed]
- Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011;124:1537-47. [Crossref] [PubMed]
- Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 2007;13:486-91. [Crossref] [PubMed]
- Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007;129:303-17. [Crossref] [PubMed]
- Kwon C, Han Z, Olson EN, et al. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 2005;102:18986-91. [Crossref] [PubMed]
- Qian L, Wythe JD, Liu J, et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol 2011;193:1181-96. [Crossref] [PubMed]
- Liu X, Zhang Y, Du W, et al. MiR-223-3p as a Novel MicroRNA Regulator of Expression of Voltage-Gated K+ Channel Kv4.2 in Acute Myocardial Infarction. Cell Physiol Biochem 2016;39:102-14. [Crossref] [PubMed]
- Wulff H, Castle NA, Pardo LA. Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov 2009;8:982-1001. [Crossref] [PubMed]
- Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014;13:622-38. [Crossref] [PubMed]
- Dangwal S, Thum T. microRNA therapeutics in cardiovascular disease models. Annu Rev Pharmacol Toxicol 2014;54:185-203. [Crossref] [PubMed]
- Qin D, Wang X, Li Y, et al. MicroRNA-223-5p and -3p Cooperatively Suppress Necroptosis in Ischemic/Reperfused Hearts. J Biol Chem 2016;291:20247-59. [Crossref] [PubMed]
- Wang X, Huang W, Yang Y, et al. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim Biophys Acta 2014;1842:701-11.
- Perrin MJ, Adler A, Green S, et al. Evaluation of genes encoding for the transient outward current (Ito) identifies the KCND2 gene as a cause of J-wave syndrome associated with sudden cardiac death. Circ Cardiovasc Genet 2014;7:782-9. [Crossref] [PubMed]
- Dixon JE, Shi W, Wang HS, et al. Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circ Res 1996;79:659-68. [Crossref] [PubMed]
- Feng J, Wang Z, Li GR, et al. Effects of class III antiarrhythmic drugs on transient outward and ultra-rapid delayed rectifier currents in human atrial myocytes. J Pharmacol Exp Ther 1997;281:384-92. [PubMed]
- Grueter CE, van Rooij E, Johnson BA, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012;149:671-83. [Crossref] [PubMed]


