The state of the art in prediction of breast cancer relapse using cell-free circulating tumor DNA liquid biopsies
In the last half-century, impressive developments in molecular and genetic techniques has provided us with powerful tools for evaluation and decision making in the diagnosis, treatment, and follow-up of breast cancer patients. Standard practice includes the use of estrogen (ER) and progesterone receptor expression levels to describe biological features and endocrine responsiveness, histological grade, Ki67, and molecular signatures to evaluate proliferation and chemotherapy sensitivity, amplification status of the oncogene HER2 to stratify patients for HER2-directed treatment, and BRCA1/BRCA2 mutation status along with other high penetrant genes for hereditary risk assessment. Still other biomarkers are under development, among which the analysis of cell-free circulating tumor DNA (ctDNA) is one of the most exciting.
There are a number of features of ctDNA that make it a promising biomarker. First, ctDNA analysis opens wide a window to the cancer genome and genotypic heterogeneity, and conceivably may give a more complete view since it is comprised of DNA released from tumor cells throughout the patient (1,2). ctDNA is also quantitative and the concentration at a given time-point appears to be generally proportional to tumor burden and tumor progression (3-5). Furthermore, “liquid biopsy” sampling is simpler, more practical, and has low risk compared to conventional tissue biopsy: ctDNA can be measured from a variety of non-invasive sources, most commonly blood plasma, but also other bodily fluids such as urine, sputum, and cerebrospinal fluid. This enables convenient repeat testing and monitoring of disease dynamics in real time.
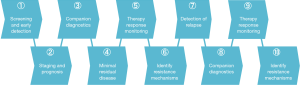
The quantitative molecular information provided by ctDNA analysis has numerous possible applications throughout the patient journey, including in the detection of cancer, staging and prognosis, companion diagnostics for actionable mutations and patient stratification, monitoring of therapy response, identification of resistance mechanisms, detection of minimal residual disease, and prediction of relapse (Figure 1). Of these, a number have already been demonstrated as feasible in breast cancer. For example, prevalent PIK3CA hotspot mutations, a target of therapeutics in development, are readily identifiable in ctDNA of patients with late as well as early breast cancer (6,7) and predict poor prognosis (8). ESR1 mutations, a mechanism of endocrine therapy resistance, can be detected by ctDNA analysis in metastatic breast cancer (MBC) (9) and associate with shorter survival (10,11). Analysis of ctDNA is more sensitive, specific, and has a greater correlation to tumor burden than antigen CA15-3 and circulating tumor cells in MBC (12). In a retrospective analysis of patients with early stage breast cancer, our group recently showed that serial ctDNA measurement could detect metastatic disease up to 37 months (average 11 months) prior to clinical detection, and that ctDNA concentration was a quantitative predictor for poor disease-free and overall survival (13). Although these studies and others demonstrate the promise of ctDNA analysis in breast cancer, most of these have had retrospective study designs. Greater evidence, in particular with prospective studies, is needed before ctDNA analyses can enter the clinical routine.
In one such landmark prospective study, Drs. Turner and Garcia-Murillas and co-workers recently described the use of single or repetitive ctDNA measurements to predict recurrence after primary breast cancer treatment in 55 early breast cancer cases treated with preoperative chemotherapy (14). Their approach was to first identify somatic mutations in the primary tumors using targeted deep sequencing of 14 genes, and then apply personalized digital PCR analyses for selected mutations in plasma samples taken at baseline (before any treatment), at 2–4 weeks after surgery, and in serial samples every 6 months postoperatively. For 43 informative patients with one or more somatic mutations identified in the primary tumor, detection of residual ctDNA [minimal residual disease (MRD)] in the first liquid biopsy after surgery, but not baseline ctDNA levels, was strongly predictive of early relapse during the follow-up period of up to three years with a hazard ratio of 25. When considering serial ctDNA measurements during postsurgical follow-up, the discriminatory value between eventual relapsing and non-relapsing patients was very high: 12 of 15 patients who relapsed had ctDNA detected in one or more post-surgical samples compared to no ctDNA detected in 27 of 28 patients who did not relapse. Using the serial ctDNA testing results, clinical recurrence could be predicted with a median lead-time of 7.9 months (range, 0.03 to 13.6 months) over clinical relapse. Importantly, the ctDNA was generally equally predictive of relapse within ER+, ER−, and triple negative disease. Also notable is that ctDNA was not detected in any of the three patients whose metastatic disease was limited to the brain, suggesting a lowered capacity for ctDNA to cross the blood-brain barrier. To further investigate whether ctDNA analysis could shed light on tumor genetic heterogeneity and evolution, five patients with MRD were further studied by targeted sequencing of tumor tissues and liquid biopsy samples for 273 genes. This revealed increased genetic diversity in 4/5 cases of MRD, including important changes such as the loss of ESR1 mutation presaging the switch from an ER+ primary to ER− metastasis in one patient. In another patient, residual disease was detected using 2 out of 3 mutants present in the primary, followed much later by the detection of a late gain of an RB1 truncating mutation. This suggests that MRD may be detected in plasma prior to the emergence of genetic diversity.
Put together, the important Garcia-Murillas et al. study adds compelling support to the idea of monitoring adjuvant treatment through serial measurements of ctDNA, and in case of positive findings, directed treatment with the purpose of eradicating residual micro-metastatic disease. Such a strategy assumes that there is a curative window of minimal disease burden in an early breast cancer patient, prior to the development and detection of clinically manifested metastatic disease, or that earlier treatment of subclinical metastatic disease can lead to meaningfully longer disease control. Using conventional imaging and serum protein markers, studies to date have not shown any benefit of monitoring for metastatic disease after primary treatment. For example, a recent Cochrane review of two randomized trials including 2,563 patients comparing conventional clinical and mammographic follow-up after primary treatment of patients with breast cancer stage I–III, versus the addition of an intense monitoring scheme of radiological and laboratory tests, did not reveal any difference in overall or disease-free survival between study groups (15). Of course, one possibility is that the imaging modalities and protein biomarkers used lacked sufficient sensitivity and/or specificity for relapse monitoring.
All evidence thus far suggests a considerably superior sensitivity and specificity of serial ctDNA analyses to detect minimally invasive disease compared to radiographic imaging and conventional laboratory analyses (12-14). Therefore, the potential benefit of early treatment in ctDNA-detected recurrent disease calls for urgent evaluation. Additional prospective and even randomized studies need to be performed and adequately powered to evaluate clinical utility within molecular subtypes, e.g., between triple negative disease with a tendency for early recurrences and luminal A cancers that can have a long subclinical period until clinical relapse. The effects of repeat disease monitoring on quality-of-life and the risk for exposing the patient to ineffective treatments will also need to be considered.
Another area where the use of ctDNA may prove beneficial for breast cancer patients, probably even sooner than adjuvant monitoring, is the monitoring of treatment in the metastatic setting. Today’s standard procedures comprise of repeated imaging, preferably using RECIST criteria, supported by conventional circulating tumor markers (16,17). The drawbacks of imaging are evident. In non-measurable disease, such as in the situation of bone-only disease, imaging is a relatively ineffective tool for disease evaluation. Patients with non-measurable disease are often excluded from clinical trials. Repeated imaging is associated with considerable side effects for the patients undergoing contrast enhanced CAT scans, and the use of more sophisticated radiotracer imaging techniques are associated with a high economic and environmental cost. Available data suggest that ctDNA-levels reflect the tumor burden more accurately compared with available circulating markers (3-5,12). In addition, the level of evidence supporting current imaging techniques is weak. These facts and the addition of companion diagnostic information on actionable mutations, with a potential to direct treatment during the metastatic course, may accelerate the introduction of ctDNA testing in the routine clinical care for metastatic patients.
More data is needed before consecutive ctDNA determinations can replace current standard procedures in clinical praxis, but it is not farfetched to anticipate a future where ctDNA-based monitoring of breast cancer treatment in the preoperative, adjuvant, and metastatic settings will prove not only cost-effective for the health care providers, but also beneficial and convenient for the patients.
Acknowledgements
This work was supported in part by the Swedish Cancer Society, Swedish Research Council, Vinnova, Region Skåne Research Funds, and the Governmental Funding of Clinical Research within National Health Service (ALF).
Footnote
Provenance: This is a Guest Commentary commissioned by Section Editor Bingrong Zhou, MD, PhD (Department of Dermatology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014;25:1729-35. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker--a critical appraisal of the literature. Clin Chim Acta 2010;411:1611-24. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014;20:2643-50. [Crossref] [PubMed]
- Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 2010;120:461-7. [Crossref] [PubMed]
- Oshiro C, Kagara N, Naoi Y, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 2015;150:299-307. [Crossref] [PubMed]
- Wang P, Bahreini A, Gyanchandani R, et al. Sensitive Detection of Mono- and Polyclonal ESR1 Mutations in Primary Tumors, Metastatic Lesions, and Cell-Free DNA of Breast Cancer Patients. Clin Cancer Res 2016;22:1130-7. [Crossref] [PubMed]
- Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2016;2:1310-5. [Crossref] [PubMed]
- Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 2015;7:313ra182. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015;7:1034-47. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- Moschetti I, Cinquini M, Lambertini M, et al. Follow-up strategies for women treated for early breast cancer. Cochrane Database Syst Rev 2016.CD001768. [PubMed]
- Cardoso F, Costa A, Norton L, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2)†. Ann Oncol 2014;25:1871-88. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]


