The role of genetics and antibodies in sepsis
Introduction
Sepsis is a heterogeneous, life threatening syndrome that can affect young and otherwise healthy people and elderly with a variety of co-morbidities. The heterogeneous nature of this disorder is better reflected by the difficulties in framing a consistent definition. Current opinion suggests that due to the existing heterogeneity sepsis definition should reflect the complex pathophysiology of sepsis. As such, sepsis is nowadays recognized as a life-threatening organ dysfunction(s) that is caused by the dysregulated response of the host to an infection (1).
The innate immune system is the first line of defense against infections yet often collaborates with adaptive immune responses to protect the host. For example, the alternative complement (C’) pathway and the mannose-binding lectin pathway are the innate elements of the complement system, which are complemented by the classical, antibody-dependent C’ system in dealing with most infectious threats. It is postulated that during the progression of sepsis the host is not able to produce an adequate amount of immunoglobulins (IGs). This generates the hypothesis that exogenous replacement of IGs may be a candidate therapy (2). However, the production of IGs is governed by the coordination of T- and B-lymphocytes whatever may differ according to the genetic make-up of the host.
This current review highlights the evidence on how the genetic diversity of the host may have an impact on the production of IGs but also how this may affect the response of the sepsis host to the exogenous administrations of antibodies. Exogenously administered antibodies may largely be either Ig of classes G, M and A or recombinant antibodies targeting specific structures of the host or the pathogens.
IGs and sepsis pathogenesis
While the exact pathophysiology of sepsis remains largely unknown, a variety of factors interfere to modulate the response of the host to an infectious stimulus and this has direct implications both on clinical severity and outcome. Using a largely simplistic scheme, it is postulated that sepsis starts after recognition of well-evolutionary conserved microbial structures called pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) of the innate immune system. The interaction between PAMPs and PRRs leads to the production and subsequent release of pro-inflammatory and anti-inflammatory mediators that orchestrate the clinical state of the host. Key effector cells in the PAMP-PRR interaction are Ag-presenting cell that through the activation of major human leukocyte antigen II (HLA) and co-stimulatory molecules like CD86 activate naïve T cells into T-helper (Th) cells either with pro-inflammatory properties (Th1 and Th17 cells) or into T cells with anti-inflammatory properties (Th2 and T regulatory cells) (3). These Th cells through the production of cytokines, mainly tumor necrosis factor-alpha (TNFα) and interleukin (IL)-6, stimulate the maturation, clonal expansion and differentiation of B-lymphocytes into plasma cells that release IGs (4). The most important IGs are the subclasses of IgG that form dimers and IgM that forms potent polyvalent pentamers with ten binding sites. IgM antibodies are the first to be produced upon recognition of an unknown Ag followed by IgG production upon generation of immune memory.
It is currently conceived that patients with sepsis can be grouped into four categories regarding the intensity of the inflammatory phenomena they experience: those with predominant pro-inflammatory responses usually found when sepsis develops in a young otherwise healthy individual; those with predominant anti-inflammatory responses usually developing in immune suppressed individuals; those with fluctuating pro- and anti-inflammatory responses usually developing in healthy individuals where the infection source is not adequately controlled; and those starting with pro-inflammatory responses followed by an anti-inflammatory course which is the typical sequence of events in the majority of patients (5). Anti-inflammation is characterized by failure of the immune system to respond adequately to a bacterial stimulus. At that time course, lymphopenia predominates part of which involves B-lymphocytes and subsequent the reduced capacity for adequate production of IGs (6).
Several recent studies suggest that circulating IGs are decreased when sepsis emerges. The first small study enrolled 21 patients with septic shock; 16 had hypoglobulinemia. These patients could be classified into those with selectively low IgG, into those with selectively low IgM and into those with combined low IgG and IgM (7). In a study of 62 patients, 61% low IgG, 40% had low IgM and 4% of patients had low IgA at the start of septic shock (8).
Two recent large studies coming from Spain analyzed immunoglobulin levels in sepsis. The first study in 172 patients was a multicenter, prospective study from nine hospitals. Concentrations of IgG, IgM and IgA were measured on the first day of severe sepsis or septic shock. Using cut-offs of 300 mg/dL for IgG1, 35 mg/dL for IgM and 150 mg/dL for IgA, all patients were classified as below or above this cut-off. Then using logistic regression analysis, three immuno-scores were described to be associated with unfavorable outcome: (I) all three IgG1, IgM and IgA below the cutoffs with odds ratio 5.27 for death; (II) both IgG1 and IgM below the cutoffs with odds ratio 3.10 for death; and (III) both IgG1 and IgA below the cutoffs with odds ratio 4.10 for death (9). A total of 171 patients with community-acquired pneumonia (CAP) were enrolled in the second multicenter study. It was found that low subclasses of IgG were mostly associated with disease severity (10).
The largest study conducted so far involved the measurements of IgM in 332 Greek patients by the Hellenic Sepsis Study Group (www.sepsis.gr). A total of 41 patients had systemic inflammatory response syndrome (SIRS) due to acute pancreatitis, 100 uncomplicated sepsis, 113 severe sepsis and 78 patients had septic shock. Circulating IgM measured within the first 24 hours was significantly decreased in uncomplicated sepsis and septic shock but not in severe sepsis in comparison with healthy controls. Repeated measurements showed decrease of IgM the first 24 hours of transition from severe sepsis into septic shock. For patients with septic shock, daily IgM measurements were done, starting from the first 24 hours from vasopressors for another 6 days. The body distribution of IgM reflected by the area under curve of IgM over the entire time of follow-up was significantly greater in survivors than in non-survivors both when censoring was done on day 28 or on the day of hospital discharge showing a great body deficit of IgM linked with unfavorable outcome (11). This deficit in the body distribution of IgM was also associated with failure of ex vivo stimulation of circulating lymphocytes by the universal lymphocyte agonist phytohemagglutin for the production of IgM.
A recent study on the transcriptional activity of peripheral blood leukocytes of patients with CAP hospitalized in an ICU comprised a discovery cohort of 265 patients and a validation cohort of 106 patients. Results showed down-regulation of pathways associated with activation of both T- and B-lymphocytes (12). These gene profiles are in keeping with our above findings of anergy of lymphocytes of sepsis patients for the production of IgM (11).
A genetic perception on pathogenesis
However, not all patients with sepsis develop a clinical condition of the same severity. This is partly due to the fact that the quantitative characteristics of the PAMPs and PRRs interaction are not similar in all patients. However, it may even be the case that the qualitative characteristic of this interaction is largely different between patients. PRRs and mediators of inflammation are mostly protein molecules encoded by genes. Gene nucleotide sequences may differ within hosts and these single nucleotide polymorphisms (SNPs) may theoretically elicit a different constellation of host-microbe interaction and impact on clinical outcome (Figure 1). According to this hypothesis, SNPs residing on each of the following structures may affect susceptibility to sepsis or sepsis outcome:
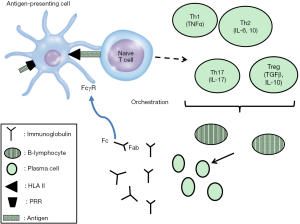
- PRRs on Ag presenting cells: PRRs, namely Toll-like receptors (TLRs) and triggering receptor expressed on myeloid cells (TREMs) are directly stimulated by microbial PAMPs ending in the production of cytokines. Some TLRs like TLR4 interact with CD14 for the binding of lipopolysaccharides (LPS) of Gram-negative bacteria. During the state of sepsis immunosuppression, the expression of HLA and co-stimulatory molecules like CD86 is decreased (13);
- Cytokines: cytokines like pro-inflammatory molecules TNFα, IL-1 beta, IL-12, type 1 and 2 interferons, and predominately anti-inflammatory cytokines like IL-4, IL-10, and transforming growth factor beta (TGFβ);
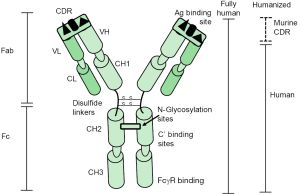
- IGs and their receptor binding sites found on the crystallizable fragment of immunoglobulin (Fc) of IG bind to receptors expressed on Ag presenting cells, phagocytes and natural killer cells. Fc gamma receptors (FcγRs) can mediate activation signals via one of five activation receptors (FcγRI, FcγRIIa, FcγRIIc, FcγRIIIa, and FcγRIIIb) or the inhibitory Fc receptor FcγRIIb. The activating Fc receptors possess an immunoreceptor tyrosine-based activation motif (ITAM) in their intracellular domain. The ITAM activates the SRC family of kinases and spleen tyrosine kinase (14). In contrast, the inhibitory FcγRIIb contains an immunoreceptor tyrosine-based inhibitory motif (ITIM) in its intracellular domain that features an activation site for the SH2 domain-containing inositol 5’ phosphatase 1 (SHIP-1) that down regulates intracellular signaling cascades (14).
Commenting on available methodologies
Research on the importance of SNPs for susceptibility to a disorder may follow three types of experimental approach: (I) case-control studies; (II) genome-wide association studies (GWAS); and (III) whole exome sequencing (WES) studies. The exome is the entire array of coding sequences for transcription and translation into protein products. Case-control studies are based on the comparison of the frequency of a candidate gene between patients having the disorder under study and matched controls. Once statistical difference is disclosed, the finding should be validated by multivariate logistic regression analysis taking into consideration disease severity and patients’ co-morbidities. Two major limitations of case-control studies in sepsis should be recognized: (I) the source of controls being in some studies healthy people and in other studies critically ill patients; and (II) the lack of consideration that protein molecules are under a dynamic interaction and their end result may be confounded by a SNP of a protein participating in the same or parallel pathway as the protein under study. To overcome this hurdle, GWAS studies are running a broad constellation of SNPs on a predefined SNP chip. The limitation of GWAS is that not all SNPs can be included in a single chip. WES takes into account the analysis of the nucleotide sequencing of the entire host exome.
The vast majority of studies in sepsis are case-association studies of candidate SNPs. Only one GWAS has been published so far on patients with CAP (15); no WES has been published so far in sepsis patients.
Modulation of sepsis pathogenesis by the genetic background
This manuscript refers to the available evidence of the biological role of SNPs of (I) PRRs on Ag-presenting cells; (II) cytokines and (III) IGs on susceptibility to sepsis and sepsis outcome. The SNPs for which most of available evidence is repetitive and robust are summarized in Table 1.

Full table
PRRs
The most broadly studied SNPs are those of the TLR4 gene encoding for TLR4 that is the receptor of LPS. The best described SNPs are rs4986790 that causes a substitution of aspartic acid by glycine at amino acid position 299 (Asp299Gly) and rs4986791 that causes the substitution of threonine by isoleucine at amino acid position 399 (Thr399Ile). In the Caucasian populations these SNP alleles exist as a haplotype on the same chromosome and they do not affect cytokine production. However, African populations bear haplotypes composed of the SNP allele at position 299 and of the wild-type allele at position 399. This haplotype is associated with high production of TNFα from monocytes after LPS stimulation (19). This is considered to be an evolutionary characteristic offering survival benefit among African populations against malaria. A meta-analysis of 17 case-association studies mostly conducted in Caucasian populations has failed to show any impact of TLR4 SNPs on susceptibility to sepsis (16). However, in Chinese Han patients one TACCCG haplotype has been described that is associated with 1.59 odds ratio for sepsis acquisition (P=0.006). This haplotype consists of rs10759932 T/C, rs11536879 A/G, rs12377632 C/T, rs1927907 C/T, rs11536889 G/C and rs7873784 G/C (20). The salient SNP in this haplotype seems to be rs11536889 since presence of only G alleles was found in Chinese patients to be associated with greater sequential organ failures assessment (SOFA) scores for renal, liver and coagulation function. However, overall survival compared to patients bearing at least one C allele remained unchanged (21).
As mentioned, prior to TLR4 binding LPS is attached to CD14. The broadest studied SNP of CD14 is rs2569190 encoding for a C/T transition at the −159 position of the gene promoter. A meta-analysis of 17 studies has shown that carriers of C alleles are exposed to 1.72 odds ratio for septic shock (P=0.03) but this association existed mainly for Asian populations and not for European populations (22). In a recent study of 417 adult patients of western European descent, 30-day survival was 77% among patients with the CT or TT genotype and 87% among patients with the TT genotype (P=0.0261) (23).
Several studies are also published on the significance of rs5743708 of TLR2 leading through one A/G transition to a substitution of arginine by glutamate at position 753. TLR2 encodes for TLR2 that is the receptor for the cell wall PAMPs of Gram-positive cocci. A meta-analysis of 12 studies has shown that carriers of G alleles are at a greater risk for sepsis acquisition. Despite the great heterogeneity of this meta-analysis, this risk association was found using both the allelic and the recessive analysis approach (24).
TLR1 participates also in monocyte activation by Gram-positive cocci. The SNP re5743661 encodes for a nonsynonymous A/G transition at position −7202. Although carriage does not increase susceptibility to sepsis among multiple trauma patients, it is associated with 4.88 odds ratio for mortality after 30-days from Gram-positive sepsis (P=0.013) (25).
A small study in Chinese Han populations has shown an impact of SNPs of TREM-1 for susceptibility to septic shock. A total of 124 patients with septic shock and 152 controls were genotyped for rs2234246 leading to one G/A transition at gene promoter. Carriage of the A allele was associated with 3.10 odds ratio for septic shock (P=0.020) and this was proved using both the recessive and the codominant analysis models (26).
As already described, the patient with sepsis is gradually progressing to a state of functional suppression of circulating monocytes. At that state, down-regulated expression of CD86 and HLA molecules on the surface of monocytes will occur. Only two studies until now have attempted to analyze the impact of SNPs of these molecules on the clinical course of sepsis. The first study enrolled 192 patients with lung infection and sepsis. Authors described a protective role from carriage of rs17281995 C allele of CD86 in patients compared to healthy controls. This was associated with lower gene expression and protein release of CD86 from monocytes (27). A haplotype of HLA-G was also studied in a prospective study of 638 patients admitted in an intensive care unit (ICU) in Rio Grande do Sul in Brazil. The haplotype consisted of an insertion at position +2960 (rs1704), of the G allele at position +3142 (rs1063320) and an insertion at position +3187 (rs9380142). Carriers of the haplotype had 1.62 odds ratio for septic shock (P=0.031) (17).
Cytokines
Several case-control studies have been published on the impact of candidate SNPs of genes encoding for cytokines on sepsis susceptibility and sepsis outcome. Here, we focus on only two genes, TNF and IL-6, encoding for TNFα and IL-6 respectively, that are an expression of Th1 and Th2 activation. The best studied SNP of TNF is rs1800629 leading to one A/G transition at −308 position of gene promoter. The significance of the carriage of this SNP had been fully clarified in one meta-analysis of 25 case-association studies (28). Results showed that carriage of A alleles was associated with susceptibility to sepsis but not with mortality. Recent publications not only study the impact of the rs1800629 SNP but they analyze further the role of other SNPs of the gene promoter. In particular, rs1800750 leading to one G/A transition at the −376 position and rs361525 leading to one G/A transition at the −238 position. Carriage of A alleles of these SNPs is associated with increased susceptibility for severe sepsis but does not predict increased lethality among Chinese Han populations (29).
The susceptibility for sepsis acquisition was shown in a prospective study of our group on 213 patients. These patients were mechanically ventilated and all developed ventilator-associated pneumonia (VAP). Carriers of A alleles for any of three promoter SNPs progressed to VAP earlier than their comparators carrying only the wild-type G alleles (30). The carriage of the TNF SNPs is associated with modulation on TNFα production by circulating monocytes. Both in our study and in others (30,31) circulating leukocytes of patients carrying these SNPs produced greater quantities of TNFα after stimulation by bacterial LPS.
The broadest studied SNP of IL-6 is rs1800795 leading to one G/C transition at the −174 promoter region. A meta-analysis of 12 case-control studies failed to demonstrate a real effect of carriage of minor frequency C alleles on sepsis risk (18). However, considerable heterogeneity exists in these studies as patients of European and African ancestry were analyzed together and the control groups were also very heterogeneous. Another study of 1,246 white Spanish patients with CAP failed to show an effect on sepsis risk; however, patients homozygous for the C minor frequency allele were protected from progression into septic shock, acute respiratory distress syndrome (ARDS), multiple organ dysfunction syndrome (MODS) and death (32).
However, SNPs of this gene seem to be a good example of the individualized impact of the genetic make-up of the host in relation to his overall health state. It is broadly known that SNPs of IL-6 are associated with the risk for cardiovascular disorders and chronic kidney disease (CKD). Since CRD is a predisposing condition for sepsis per se it is difficult to discriminate an effect from IL-6 SNPs on sepsis susceptibility versus the SNP impact from CKD. We analyzed rs1800795 and a second SNP rs1800796 encoding for one G/C transition at the −572 promoter region among 198 sepsis patients with CKD; 115 patients with CKD without infection were used as controls. The presence of minor frequency alleles of rs1800795 did not affect sepsis risk. However, carriage of the GG genotype of rs1800796 was associated with 2.07 odds ratio for sepsis (P=0.005). It was interesting to note that despite this difference in susceptibility for sepsis, no overall effect on mortality was shown. Sub-analysis revealed that patients with the GG genotype were at increased risk for death by cardiovascular events whereas patients with the GC and CC genotype were at increased risk for sepsis-related death (33).
IGs
When focusing on the concentrations of circulating antibodies in patients with sepsis, it becomes evident that they follow linear distribution. Comparisons between groups based upon disease are done with non-parametric statistics (8-11). This means that there is an individuality for IG production within each group. This generates the notion that the genetic background of the host may play a major role in the capacity for IG production irrespective of sepsis severity.
Only three studies on SNPs of genes encoding for antibody peptides are available. All three report on rs1801274 of CD32 encoding for the activating receptor of the FcγRIIa. The SNP leads to one G/A transition and a substitution of arginine by histidine at position 131. It is anticipated that this SNP decreases the binding affinity of the FcγRIIa and decreases phagocytic activity of neutrophils impairing antibody-dependent cellular phagocytosis (ADCP). The first study enrolled 347 sepsis patients using 171 non-infected critically ill patients as comparators. Carriage of SNP alleles was not associated with sepsis risk (34). The second study enrolled 66 ICU-admitted patients for multiple injuries; 36 developed sepsis and 30 were comparators. Presence of at least one A allele was associated with increased likelihood for sepsis (hazard ratio 3.77) even after adjustment by the Injury Severity Index. GA heterozygotes or AA homozygotes had a greater degree of monocyte deactivation as expressed by the expression of HLA-DR on circulating monocytes. The monocytes of patients showed down-regulation for the production of TNFα following ex vivo stimulation with bacterial LPS. This phenomenon was more pronounced among carriers of the wild-type GG genotype than among patients with the GA or AA genotypes (35). The last study genotyped 117 patients with pneumococcal sepsis and failed to identify some impact of SNP carriage on final outcome (36).
Treatment with antibodies and the genetic make-up of the host
Antibodies used as adjunctive therapy of sepsis are divided into two categories regarding their mode of action: (I) recombinant or chimeric molecules aiming to block specific targets; and (II) intravenous preparations of IGs either containing only IgG immunoglobulins (IVIG) or enriched in IgM and IgA (IgGAM).
Several phase 3 randomized clinical trials have been conducted in the past, mostly studying the efficacy of antibodies targeting TNFα on sepsis outcome. Although none of them proved clinical efficacy in reduction of 28-day mortality (37), none of these trials did genotyping for SNPs of the TNF gene loci. The importance of genotyping to unmask the real efficacy of this therapeutic approach is recently revealed using as a prototype hidradenitis suppurativa (HS) that is a chronic inflammatory skin disorder. HS is characterized by heavy inflammation of skin areas rich in apocrine glands. Contrary to other chronic inflammatory conditions, the abundance of TNFα in the affected lesions is accompanied by reciprocal down-regulation of circulating monocytes for cytokine production (38), providing similarities with sepsis immunosuppression. Adalimumab, a fully human, recombinant anti-TNF antibody, has recently proved clinical efficacy in two randomized phase 3 clinical trials and it is registered since 2015 for the management of HS (39). When patients under treatment with agents blocking TNFα were genotyped for the haplotypes of rs1800629, rs1800750 and rs361525 of the promoter, it was shown that the carriage of at least one minor frequency SNP allele in any of the three positions was associated with 2.67 odds ratio for reduced treatment efficacy (40). Since the carriage of SNP haplotypes in the general population exceeds 20%, it is evident that the lacking efficacy in sepsis trials may be confounded by their presence.
The use of IVIG is not recommended in sepsis patients according to the 2012 recommendations of the Surviving Sepsis Campaign (41). However, in the same recommendations it is suggested that the clinical utility of IgGAM is still an open question and its benefit should be validated in future clinical trials. A meta-analysis of the efficacy of IG preparations in sepsis has evaluated the efficacy of administration of IgGAM in a total of 528 adults enrolled in seven trials. The authors concluded that treatment with IgGAM considerably decreased the risk for death after 28 days in both adults and neonates with severe sepsis and/or septic shock (odds ratio 0.66) (42).
In light of the ambiguities for the clinical efficacy of IgGAM generated by the limited number of patients enrolled in the conducted clinical trials, the Hellenic Sepsis Study Group adopted a retrospective approach. One hundred patients with severe documented infections by multidrug-resistant (MDR) Gram-negative bacteria were treated with IgGAM within the first 24 hours from the first organ failure. A total of 100 untreated historical controls were selected from the available database. Control patients were 1:1 matched for severity, MDR pathogens and appropriateness of administered antimicrobials and matched for the source of infection and Charlson’s comorbidity index. Mortality after 28 days was 58% among the comparators and 39% among patients treated with IgGAM (P=0.009). Treatment with IgGAM was associated with significant delay for breakthrough bacteremia compared to comparators (P<0.0001) (43). The aforementioned analysis of genetic SNPs affecting the cascade ending to IG production generates several questions as to whether clinical efficacy from IgGAM treatment should be adjusted by a genomic analysis encompassing all analyzed SNPs.
The use of monoclonal antibodies (mAbs) in the treatment of bacterial sepsis
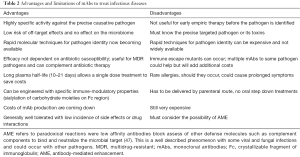
While pooled immunoglobulin preparations have been studied for decades in sepsis, the use of mAbs targeting the pathogen responsible for inducing sepsis has only recently been investigated. When Kohler and Milstein first succeeded in generating functional antibodies against specific antigenic targets, they revolutionized the field of immunology and paved the way for therapeutic mAbs for human disease (44). These highly specific antibodies have been widely used in neoplastic diseases, immune-mediated diseases and in organ transplantation, but have received, for a variety of reasons, very little attention for the management of infectious diseases. This situation is rapidly changing as the era of progressive emergence of antibiotic resistance is now upon us (45-47). IGs have much to be desired characteristics and have proved to be remarkably flexible, stable and readily engineered to alter and improve their activity (47). Figure 2 depicts immunoglobulin G and its major structural and functional components. Table 2 outlines the favorable characteristics and potential limitations of mAbs as therapeutic agents to treat infectious diseases (47-52). Additionally, the costs for developing therapeutic mAbs have previously been thought to be cost prohibitive for clinical use. Recent improvements in rapid microbial diagnosis using molecular mechanisms and reduced cost in manufacture of modern mAbs has made this field of therapeutics a more realistic approach to managing common infections.
Full table
MAbs can now be directed against bacterial infections by at least three different strategies: (I) bacterial exotoxins and other virulence factors such as biofilm synthesis; (II) promote phagocytosis and bacteriolysis by either natural killer cell mediated, antibody-dependent cellular cytotoxicity (ADCC), neutrophil mediated, antibody-dependent cellular phagocytosis (ADCP), or by facilitating C’ dependent cytotoxicity (CDC); or (III) targeting excessive or deleterious host inflammatory mediators, clearance of pathogen-derived toxic bio lipids, or blockade of endogenous vasoactive peptides such as adrenomedullin (48-51).
The specific bacterial pathogens that are currently being targeted with mAbs are Staphylococcus aureus (S. aureus), Staphylococcus epidermidis, and MDR strains of Pseudomonas aeruginosa, Escherichia coli and Klebsiella pneumoniae. The presence of hundreds of different serotypes of MDR Gram-negative enteric pathogens does not seem to be an attractive target for therapeutic mAbs. However, a relatively small number of specific clones of MDR pathogens are circulating at the present time such as E. coli ST131 O25b:H4 (52), and K. pneumoniae ST258 D-galactan III (53) are responsible for significant proportion of human infections. Antibodies are in development that could complement concurrent antimicrobial therapy or provide some protection for even pan-resistant genetic variants of these bacterial clones.
Pseudomonas aeruginosa has been a potential target for mAb immunotherapy for decades. Two approaches are currently underway with either serotype-specific, opsonic antibody approaches; and a more complex bispecific mAb construct simultaneously directed to a type 3 secretion system for cytotoxin delivery and an enzyme essential for exosaccharide-dependent biofilm formation (54,55).
There are many molecular targets amenable to monoclonal antibody therapy for S. aureus and to a lesser degree for S. epidermidis, where the primary issue with coagulase staphylococci is biofilm formation on vascular catheters and prosthetic devices (56). Prominent among these targets against S. aureus is alpha toxin and a number of other related two component leukotoxins (57-59). Other targets are various adhesions and inhibition of biofilm formation (60). Staphylococcal alpha toxin is an attractive target as this is a potent cytotoxin that targets macrophages and impairs phagocytosis. A monoclonal antibody to alpha toxin has recently been shown to protect against mixed infection murine lung models consisting of both S. aureus and K. pneumoniae (61). Prevention of phagocytic damage by alpha toxin repairs clearance of both S. aureus and other respiratory co-pathogens such as Klebsiella spp. that might be encountered in VAP.
Another innovative potential approach to the use of mAbs is the use of non- neutralizing mAbs that can affect the host response by manipulation of the Fc:FcγR interactions (62). FcγRIIb is an inhibitory Fc receptor with an intracellular ITIM domain (14). Sialyation of the glycosylated regions linked to the asparagine residue at position 297 of the Fc component of IgG can promote FcγRIIb expression and down regulate myeloid cells (63). This strategy could allow fine tuning of the host immune system for certain infections and inflammatory states.
Conclusions
This review makes it evident that there is lack of clear-cut evidence whether the genetic background of the host modulates endogenous IGs production or impacts on response to exogenous antibody administration. Given the proven down-regulation of B-lymphocyte responses in sepsis, and with the recent understanding of the feedback interactions between IGs and the innate immune effector cells, a genomic approach should take into consideration the SNPs of all molecules implicated in the pathway from Ag presentation to immunoglobulin production.
Acknowledgements
None.
Footnote
Conflicts of Interest: Evangelos J. Giamarellos-Bourboulis has received honoraria (paid to the University of Athens) from AbbVie, Biotest, Brahms GmbH, and The Medicines Company; has received compensation as a consultant for Astellas Greece (paid to the University of Athens); and has received independent educational grants (paid to the University of Athens) from AbbVie, Biotest, Brahms GmbH, The Medicines Company and Sanofi. He is funded by the FrameWork 7 program HemoSpec (granted to the University of Athens). Steven M. Opal is a consultant to Arsanis, Aridis, Biotest, Cyon, SOBI and Battelle and his institution receives grant support from Asahi-Kasei, Ferring and Biocartis.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Giamarellos-Bourboulis EJ. What is the pathophysiology of the septic host upon admission? Int J Antimicrob Agents 2010;36 Suppl 2:S2-5. [Crossref] [PubMed]
- Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol 2010;40:1830-5. [Crossref] [PubMed]
- Rauch PJ, Chudnovskiy A, Robbins CS, et al. Innate response activator B cells protect against microbial sepsis. Science 2012;335:597-601. [Crossref] [PubMed]
- Reinhart K, Bauer M, Riedemann NC, et al. New approaches to sepsis: molecular diagnostics and biomarkers. Clin Microbiol Rev 2012;25:609-34. [Crossref] [PubMed]
- Boomer JS, To K, Chang KC, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 2011;306:2594-605. [Crossref] [PubMed]
- Taccone FS, Stordeur P, De Backer D, et al. Gamma-globulin levels in patients with community-acquired septic shock. Shock 2009;32:379-85. [Crossref] [PubMed]
- Venet F, Gebeile R, Bancel J, et al. Assessment of plasmatic immunoglobulin G, A and M levels in septic shock patients. Int Immunopharmacol 2011;11:2086-90. [Crossref] [PubMed]
- Bermejo-Martín JF, Rodriguez-Fernandez A, Herrán-Monge R, et al. Immunoglobulins IgG1, IgM and IgA: a synergistic team influencing survival in sepsis. J Intern Med 2014;276:404-12. [Crossref] [PubMed]
- de la Torre MC, Bolíbar I, Vendrell M, et al. Serum immunoglobulins in the infected and convalescent phases in community-acquired pneumonia. Respir Med 2013;107:2038-45. [Crossref] [PubMed]
- Giamarellos-Bourboulis EJ, Apostolidou E, Lada M, et al. Kinetics of circulating immunoglobulin M in sepsis: relationship with final outcome. Crit Care 2013;17:R247. [Crossref] [PubMed]
- Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 2016;4:259-71. [Crossref] [PubMed]
- Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis 2013;13:260-8. [Crossref] [PubMed]
- Guilliams M, Bruhns P, Saeys Y, et al. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol 2014;14:94-108. [Crossref] [PubMed]
- Rautanen A, Mills TC, Gordon AC, et al. Genome-wide association study of survival from sepsis due to pneumonia: an observational cohort study. Lancet Respir Med 2015;3:53-60. [Crossref] [PubMed]
- Zhu L, Li X, Miao C. Lack of association between TLR4 Asp299Gly and Thr399Ile polymorphisms and sepsis susceptibility: a meta-analysis. Gene 2012;501:213-8. [Crossref] [PubMed]
- Graebin P, Veit TD, Alho CS, et al. Polymorphic variants in exon 8 at the 3' UTR of the HLA-G gene are associated with septic shock in critically ill patients. Crit Care 2012;16:R211. [Crossref] [PubMed]
- Gao JW, Zhang AQ, Pan W, et al. Association between IL-6-174G/C polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One 2015;10:e0118843. [Crossref] [PubMed]
- Ferwerda B, McCall MB, Alonso S, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci U S A 2007;104:16645-50. [Crossref] [PubMed]
- Wang H, Wei Y, Zeng Y, et al. The association of polymorphisms of TLR4 and CD14 genes with susceptibility to sepsis in a Chinese population. BMC Med Genet 2014;15:123. [Crossref] [PubMed]
- Mansur A, von Gruben L, Popov AF, et al. The regulatory toll-like receptor 4 genetic polymorphism rs11536889 is associated with renal, coagulation and hepatic organ failure in sepsis patients. J Transl Med 2014;12:177. [Crossref] [PubMed]
- Zhang AQ, Yue CL, Gu W, et al. Association between CD14 promoter -159C/T polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One 2013;8:e71237. [Crossref] [PubMed]
- Mansur A, Liese B, Steinau M, et al. The CD14 rs2569190 TT Genotype Is Associated with an Improved 30-Day Survival in Patients with Sepsis: A Prospective Observational Cohort Study. PLoS One 2015;10:e0127761. [Crossref] [PubMed]
- Gao JW, Zhang AQ, Wang X, et al. Association between the TLR2 Arg753Gln polymorphism and the risk of sepsis: a meta-analysis. Crit Care 2015;19:416. [Crossref] [PubMed]
- Thompson CM, Holden TD, Rona G, et al. Toll-like receptor 1 polymorphisms and associated outcomes in sepsis after traumatic injury: a candidate gene association study. Ann Surg 2014;259:179-85. [Crossref] [PubMed]
- Peng LS, Li J, Zhou GS, et al. Relationships between genetic polymorphisms of triggering receptor expressed on myeloid cells-1 and septic shock in a Chinese Han population. World J Emerg Med 2015;6:123-30. [Crossref] [PubMed]
- Song H, Tang L, Xu M, et al. CD86 polymorphism affects pneumonia-induced sepsis by decreasing gene expression in monocytes. Inflammation 2015;38:879-85. [Crossref] [PubMed]
- Teuffel O, Ethier MC, Beyene J, et al. Association between tumor necrosis factor-alpha promoter -308 A/G polymorphism and susceptibility to sepsis and sepsis mortality: a systematic review and meta-analysis. Crit Care Med 2010;38:276-82. [Crossref] [PubMed]
- Song Z, Song Y, Yin J, et al. Genetic variation in the TNF gene is associated with susceptibility to severe sepsis, but not with mortality. PLoS One 2012;7:e46113. [Crossref] [PubMed]
- Kotsaki A, Raftogiannis M, Routsi C, et al. Genetic polymorphisms within tumor necrosis factor gene promoter region: a role for susceptibility to ventilator-associated pneumonia. Cytokine 2012;59:358-63. [Crossref] [PubMed]
- Kothari N, Bogra J, Abbas H, et al. Tumor necrosis factor gene polymorphism results in high TNF level in sepsis and septic shock. Cytokine 2013;61:676-81. [Crossref] [PubMed]
- Martín-Loeches I, Solé-Violán J, Rodríguez de Castro F, et al. Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intensive Care Med 2012;38:256-62. [Crossref] [PubMed]
- Panayides A, Ioakeimidou A, Karamouzos V, et al. -572 G/C single nucleotide polymorphism of interleukin-6 and sepsis predisposition in chronic renal disease. Eur J Clin Microbiol Infect Dis 2015;34:2439-46. [Crossref] [PubMed]
- Beppler J, Koehler-Santos P, Pasqualim G, et al. Fc Gamma Receptor IIA (CD32A) R131 Polymorphism as a Marker of Genetic Susceptibility to Sepsis. Inflammation 2016;39:518-25. [Crossref] [PubMed]
- West SD, Ziegler A, Brooks T, et al. An FcγRIIa polymorphism with decreased C-reactive protein binding is associated with sepsis and decreased monocyte HLA-DR expression in trauma patients. J Trauma Acute Care Surg 2015;79:773-81. [Crossref] [PubMed]
- Garnacho-Montero J, García-Cabrera E, Jiménez-Álvarez R, et al. Genetic variants of the MBL2 gene are associated with mortality in pneumococcal sepsis. Diagn Microbiol Infect Dis 2012;73:39-44. [Crossref] [PubMed]
- Kotsaki A, Giamarellos-Bourboulis EJ. Emerging drugs for the treatment of sepsis. Expert Opin Emerg Drugs 2012;17:379-91. [Crossref] [PubMed]
- Kanni T, Tzanetakou V, Savva A, et al. Compartmentalized Cytokine Responses in Hidradenitis Suppurativa. PLoS One 2015;10:e0130522. [Crossref] [PubMed]
- Kimball AB, Okun MM, Williams DA, et al. Two Phase 3 Trials of Adalimumab for Hidradenitis Suppurativa. N Engl J Med 2016;375:422-34. [Crossref] [PubMed]
- Savva A, Kanni T, Damoraki G, et al. Impact of Toll-like receptor-4 and tumour necrosis factor gene polymorphisms in patients with hidradenitis suppurativa. Br J Dermatol 2013;168:311-7. [Crossref] [PubMed]
- Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 2013;41:580-637. [Crossref] [PubMed]
- Kreymann KG, de Heer G, Nierhaus A, et al. Use of polyclonal immunoglobulins as adjunctive therapy for sepsis or septic shock. Crit Care Med 2007;35:2677-85. [Crossref] [PubMed]
- Giamarellos-Bourboulis EJ, Tziolos N, Routsi C, et al. Improving outcomes of severe infections by multidrug-resistant pathogens with polyclonal IgM-enriched immunoglobulins. Clin Microbiol Infect 2016;22:499-506. [Crossref] [PubMed]
- Alkan SS. Monoclonal antibodies: the story of a discovery that revolutionized science and medicine. Nat Rev Immunol 2004;4:153-6. [Crossref] [PubMed]
- Desoubeaux G, Daguet A, Watier H. Therapeutic antibodies and infectious diseases, Tours, France, November 20-22, 2012. MAbs 2013;5:626-32. [Crossref] [PubMed]
- Casadevall A, Dadachova E, Pirofski LA. Passive antibody therapy for infectious diseases. Nat Rev Microbiol 2004;2:695-703. [Crossref] [PubMed]
- Guzman MG, Vazquez S. The complexity of antibody-dependent enhancement of dengue virus infection. Viruses 2010;2:2649-62. [Crossref] [PubMed]
- Irani V, Guy AJ, Andrew D, et al. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol Immunol 2015;67:171-82. [Crossref] [PubMed]
- Henry Dunand CJ, Leon PE, Huang M, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016;19:800-13. [Crossref] [PubMed]
- An Z, Forrest G, Moore R, et al. IgG2m4, an engineered antibody isotype with reduced Fc function. MAbs 2009;1:572-9. [Crossref] [PubMed]
- Kox M, Pickkers P. Adrenomedullin: its double-edged sword during sepsis slices yet again. Intensive Care Med Exp 2014;2:1. [Crossref] [PubMed]
- Walley KR, Francis GA, Opal SM, et al. The Central Role of Proprotein Convertase Subtilisin/Kexin Type 9 in Septic Pathogen Lipid Transport and Clearance. Am J Respir Crit Care Med 2015;192:1275-86. [Crossref] [PubMed]
- Szijártó V, Guachalla LM, Visram ZC, et al. Bactericidal monoclonal antibodies specific to the lipopolysaccharide O antigen from multidrug-resistant Escherichia coli clone ST131-O25b:H4 elicit protection in mice. Antimicrob Agents Chemother 2015;59:3109-16. [Crossref] [PubMed]
- Szijártó V, Guachalla LM, Hartl K, et al. Both clades of the epidemic KPC-producing Klebsiella pneumoniae clone ST258 share a modified galactan O-antigen type. Int J Med Microbiol 2016;306:89-98. [Crossref] [PubMed]
- Haq IJ, Gardner A, Brodlie M. A multifunctional bispecific antibody against Pseudomonas aeruginosa as a potential therapeutic strategy. Ann Transl Med 2016;4:12. [PubMed]
- Lazar H, Horn MP, Zuercher AW, et al. Pharmacokinetics and safety profile of the human anti-Pseudomonas aeruginosa serotype O11 immunoglobulin M monoclonal antibody KBPA-101 in healthy volunteers. Antimicrob Agents Chemother 2009;53:3442-6. [Crossref] [PubMed]
- Lam H, Kesselly A, Stegalkina S, et al. Antibodies to PhnD inhibit staphylococcal biofilms. Infect Immun 2014;82:3764-74. [Crossref] [PubMed]
- Hilliard JJ, Datta V, Tkaczyk C, et al. Anti-alpha-toxin monoclonal antibody and antibiotic combination therapy improves disease outcome and accelerates healing in a Staphylococcus aureus dermonecrosis model. Antimicrob Agents Chemother 2015;59:299-309. [Crossref] [PubMed]
- Poojary NS, Ramlal S, Urs RM, et al. Application of monoclonal antibodies generated against Panton-Valentine Leukocidin (PVL-S) toxin for specific identification of community acquired methicillin resistance Staphylococcus aureus. Microbiol Res 2014;169:924-30. [Crossref] [PubMed]
- Badarau A, Rouha H, Malafa S, et al. Structure-function analysis of heterodimer formation, oligomerization, and receptor binding of the Staphylococcus aureus bi-component toxin LukGH. J Biol Chem 2015;290:142-56. [Crossref] [PubMed]
- Otto M. Novel targeted immunotherapy approaches for staphylococcal infection. Expert Opin Biol Ther 2010;10:1049-59. [Crossref] [PubMed]
- Cohen TS, Hilliard JJ, Jones-Nelson O, et al. Staphylococcus aureus α toxin potentiates opportunistic bacterial lung infections. Sci Transl Med 2016;8:329ra31. [Crossref] [PubMed]
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 2006;313:670-3. [Crossref] [PubMed]


