The significance of genetics for cholangiocarcinoma development
Introduction
Cholangiocarcinoma (CCA) is a rare malignant cancer arising from cholangiocytes, the epithelial cells lining the bile ducts (1). Anatomically, CCA is classically divided in intrahepatic or extrahepatic. The intrahepatic form arises within the liver parenchyma, and the extrahepatic variant may be further subdivided in perihilar (also called Klatskin tumor) or distal tumor, with the landmark at the insertion of the cystic duct. The extent of the perihilar CCA may be described according to the Bismuth-Corlette classification (1,2).
Symptoms of CCA are often nonspecific and appear late in the course of the disease; therefore, extrahepatic cancer may show signs and symptoms related to cholestasis, such as jaundice without pain, pale stools, dark urine and pruritus, whereas intrahepatic CCA is often an incidental hepatic lesion (3,4).
To date, no specific CCA markers have been found. However, CA 19-9 (i.e. carbohydrate antigen 19-9) and CEA; (i.e. carcinoembryonic antigen) usually support the diagnosis in association with clinical, radiologic, and endoscopic findings (5-7). Despite not being specific for CCA, classical cholestatic serum parameters are often increased.
The therapeutic options for this cancer are very limited. CCA is characterized by high chemoresistance and is usually late diagnosed, providing few possibilities for surgery. These features result in low survival rates: about 50% of patients who did not receive surgery die in 3-4 months after diagnosis due to liver failure or infectious complications associated with the progressive biliary obstruction (8). On the other hand, survival rates after five-years liver resection were 20-32% for intrahepatic, 30-42% for perihilar, and 18-54% for distal CCA (9).
The election of the adequate surgery procedure is often complex and depends on the tumor stage and localization. In general, the liver resection size for intrahepatic and perihilar CCA is driven by the histological pattern and usually needs partial hepatectomy in order to achieve negative resection margins, which correlates to a better survival. On the other hand, pancreatoduodenectomy (also called Whipple resection) is indicated for distal CCA (10-12). So far, the beneficial effects of orthotopic liver transplantation (OLT) have not been completely established in terms of survival improvement compared to partial hepatectomy. However, cases of liver transplantation in selected conditions have shown promising results with regards to survival improvement (11).
CCA is usually not affected by common chemotherapies. Several studies using monotherapy or drugs combination have been performed but currently none of the antineoplastic regimens show a sufficient efficacy in CCA (13).
When surgery is not indicated, the treatment is palliative and mainly aims to reduce the biliary obstructions and infection, as well as the relative symptoms (8).
Epidemiology of CCA
Although CCA is overall a rare neoplasm accounting for 3% of all gastrointestinal tumors worldwide (14), it is the second most common primary hepatic neoplasm, after hepatocellular carcinoma (15). Several studies have reported that while the incidence and mortality rates for intrahepatic CCA are increasing worldwide, a slight decrease or stabilization for extrahepatic CCA might be occurring. In particular, the age-adjusted annual incidence of intrahepatic CCA appears to have a progressive increase in USA, from 0.13 per 100,000 persons in 1973 to 0.67 in 1997 (16) and to 0.85 during the period from 1995 to 1999 (17). In contrast, the age-adjusted incidence of extrahepatic CCA decreased from 1.08 per 100,000 in 1979 to 0.82 in 1998. Moreover, comparable trends have been shown also in the United Kingdom (18,19) and Germany (20), whereas in Italy increasing incidences for both intra and extrahepatic CCA have been reported (21). By contrast, the incidence trends in Denmark and France seem to be declining (22,23). However, over the past few years, some authors have started to investigate a number of biases that might have influenced the results of former studies on the epidemiology of CCA (24,25). The lack of a uniform classification of the heterogeneous group of CCA, the unification of biliary malignancies and other hepatocellular neoplasms (such as hepatocellular carcinoma and gallbladder cancer) in most cancer registries, and the frequent misclassification due to diagnosis in advanced stage and histological heterogeneity, are critical issues affecting not only epidemiologic studies but also the understanding of the pathophysiology of the disease (8,15).
Despite these possible controversies, a slight male preponderance and possibly differences between races are generally acknowledged (17,26). Moreover, a clear relative difference between the incidence in Eastern and Western countries is well established (17). The highest incidence rates are observed in Eastern and South-Eastern Asia, with a peak registered in Thailand [33.4 per 100,000 in men, and 12.3 per 100,000 in women, with important differences within the country itself (27,28)]. In these regions, a plain association between the infection with Opisthorchis viverrini and the development of CCA has been demonstrated (28).
Several risk factors have been extensively studied and associated with the development of CCA, such as, primary sclerosing cholangitis (PSC), liver fluke infection, hepatolithiasis or biliary malformations (4), however, the majority of patients do not develop any of these features. In addition, other risk factors such as genetic polymorphisms and life style might also contribute (26,29,30), although further studies are eagerly awaited.
Genetic alterations in cancer
Carcinogenesis is considered a multistage process that causes the malignant transformation of cells (31). Most of the gene mutations are somatic and occur as sporadic events; conversely hereditary cancer, which results from mutations inherited from parents, is less common (32,33). Up to 90% of somatic mutations are dominant, whereas only 10% of the tumors need both alleles mutation to induce tumorigenesis (33). Mutations can target the genome by changing a single nucleotides [i.e. the so called “point mutations or single nucleotide polymorphism (SNP)]”, or by altering more nucleotides, thus resulting in deletions, insertions, translocations or amplifications (34). Although mutations may occur as sporadic or inherited events, the targeted genes may be classified in: (I) oncogenes; (II) tumor suppressor; or (III) stability genes (35,36).
Mutations in oncogenes, which in physiological conditions participate in several intracellular pathways, result in their aberrant activation and therefore in loss of cell proliferation control (37).
Oncogenes-related products consist of a wide class of proteins such as transcription factors, growth factors and their receptors, signal transducers, and apoptosis regulators (35,37). Transcription factors modulate the expression of genes involved in signaling pathways via downregulation or upregulation of their transcription. For example, mutations of Fos/Jun/AP1 are detected in lymphoid cancers as Hodgkin lymphoma (38).
ERBB receptors and c-MET are both members of the growth factor receptors; the binding of specific ligands initiates intracellular cascades via tyrosine kinase autophosphorylation resulting in cell proliferation, decreased apoptosis, enhanced cancer cell motility, and regulating cell differentiation (39-42). Overexpression of ERBB receptors in several tumors is the rational to treat these cancers with drugs that inhibit tyrosine kinase activity (40,43). Among the signal transducers, K-ras mutations are widely detected in a variety of tumors such as colon cancer, pancreatic cancer, and melanoma (44). Finally, oncogenes can modify the antiapoptotic activity of some molecule as Bcl-2; aberrant activation might be thus correlated to excessive proliferation as, for example, in diffuse large B-cell lymphoma (45).
Tumor suppressor genes (TSGs) are typically recessive genes; both alleles need to be mutated in order to induce tumorigenesis, according to the so-called “two hit hypothesis” (46). Many human cancers, such as retinoblastoma and familial adenomatous polyposis (FAP), have been associated with inactivation of TSGs (47). In this regard, p53 is a fundamental regulator of the cell cycle that in case of DNA damage blocks the cell cycle and leads to cellular apoptosis (46,48).
Moreover, there is a class of cancer genes called “stability genes” composed by the mismatch repair (MMR), the nucleotide-excision repair (NER) and the base-excision repair (BER) genes. The role of these genes is to correct mismatches of bases generated during normal DNA replication or induced by mutagens. Alterations of MMR genes can induce mistakes during the DNA replication; slipped strand mispairing mutations lead to different length in DNA regions and since that condition facilitates gene mutation is called microsatellite instability (49,50).
The predisposition to develop HPCC is due to mutations in members of MMR genes as MLH1, MSH2, MSH6, and PMS2 (51).
Epigenetic alterations in cancer
The research of the last decade has highlighted that human cancers also harbor a number of other heritable abnormalities in gene expression that are not caused by mutation in any region of the genome, termed epigenetic changes (52). The most studied epigenetic changes that occur in cancer comprise DNA methylation and histone modification and, broadly, include non-coding RNAs.
Methylation of the bases that constitute the genome plays a key role in a variety of physiologic processes such as embryologic development (53), genomic imprinting (54), inactivation of the X-chromosome in females (55), and preventing DNA instability caused by transposable DNA sequences (56).
DNA methylation takes place in mammals when a methyl group is added to the cytosine that directly precedes a guanine in the genome (also called CpG site, for Cytosine-phosphate-Guanine). CpG sites are not randomly distributed throughout the genome. Indeed, stretches of CpGs, termed CpG islands, can be found in many genes at the 5' end, which corresponds to their promoter region (57). These regions are typically not methylated in normal conditions, but become hypermetylated in TSGs genes in a broad variety of tumors (52,58,59). As a result of promoter hypermethylation, the gene transcription is silenced or downregulated, and thus epigenetic changes can influence the carcinogenetic process in a similar fashion to genetic mutations. The list of TSGs found to be hypermethylated in cancer is wide and constantly growing; well-known examples are VHL in renal carcinoma (60), p16INK4a in many cancers (61), and hMLH1 in colorectal carcinoma (61). Although hypermethylation of CpG islands appears to be a major event in many cancers, hypomethylation of CpG sites is also described for many tumors (62).
An alternative epigenetic change that occurs in cancer is histone modification (63). Histones are alkaline proteins that serve as scaffolds around which DNA winds in structures called nucleosomes (64). Post-transcriptional modifications, such as acetylation, methylation and phosphorylation are common events that regulate the biology of histones. In this context, the acetylation by histone acetyltransferases (HATs) of lysine residues and the deacetylation by histone deacetylases (HDACs) are the most prominent modifications influencing histone function, and the balance between the two processes regulates, at least in part, the gene expression. Indeed, the removal of the acetyl group by HDAC leads to chromatin condensation and inhibition of transcription of the involved gene, whereas the action of HATs favors gene transcription, possibly via a more favorable DNA conformation for the binding of RNA polymerases and transcription factors (65,66).
Non-coding RNAs (ncRNAs) are a group of endogenously transcribed RNA molecules that are not translated into proteins. The large family of ncRNAs comprises different members generally divided in two major subgroups: small ncRNAs and long ncRNAs (67). In this manuscript, we will only highlight microRNAs [for a comprehensive review on ncRNA see Esteller et al., Knowling et al. (68,69)].
MicroRNAs are small RNA sequences (19 to 25 nucleotides) that are involved in many biological processes such as embryonic development, proliferation, differentiation, and cell death (70). MicroRNAs are encoded in the genome, transcribed into precursor transcripts, and undergo a series of tightly regulated processes leading to their incorporation in the RNA-inducing silencing complex (RISC). RISC then directs the modulation of mRNAs translation by the binding of the microRNA to the 3' untranslated region of the target mRNA through a partial or complete sequence homology; as a result, the translation of the mRNA may be downregulated or blocked, respectively (71). MicroRNAs have been linked to many aspects of cancer, from initiation and progression of tumors to response to therapy, and development of new treatment (72).
Genetic alterations in CCA
The specific mechanisms that occur during biliary carcinogenesis are still unclear. However, chronic inflammation, partial bile flow obstruction (i.e. cholestasis), and bile duct injury are recognized to be major features for malignant transformation (1,13).
Chronic inflammation induces the secretion of pro-inflammatory cytokines from both cholangiocytes and inflammatory cells (73). Interleukin (IL)-6 and other mediators such as endotoxins and tumor necrosis factor (TNF)-α are important cytokines produced during inflammation (74). IL-6 can activate different pathways leading to mitogenic responses and cell survival (75). IL-6 is also able to induce nitric oxide synthase (iNOS) expression, which in turn increases nitric oxide (NO) production resulting in DNA damage (76,77). In addition, such inflammatory scenario can also lead to cyclic oxygenase (COX)-2 activation, the enzyme involved in prostaglandin secretion. Bile acids and other bile components have been associated to COX-2 overexpression, resulting in cell growth, anti-apoptosis and angiogenesis (78,79).
To date, many genes have been related to cholangiocarcinogenesis (Table 1) (88). However, the specific mechanisms responsible for tumorigenesis in CCA are still under investigation. Among the growth factor receptor family, c-MET mutation was reported to frequently occur in bile duct cancer, event that correlates with high grade of invasiveness and a poor prognosis (84,89,90). On the other hand, gain-of-function mutations in ERBB2 and EGFR genes are frequently observed in several heterogeneous tumors such as breast, lung, and colon cancers (91). In this regard, EGFR overexpression correlates with malignancy in human cholangiocytes since such mutation has been detected in both gallbladder and bile duct tumors but not in physiological conditions (82,92). Similarly to EGFR, ERBB2 overexpression has also reported in CCA (83,93). The simultaneous expression of ERBB2 and COX-2 may indicate a prostaglandins secretion induced by ERBB2, which is known to be strongly mitogenic (94). Moreover, the correlation between ERBB2 mutations and tumor progression is suggested by the fact that rat cells transfected with the ERBB2/neu oncogene show features similar to human CCA (95). In terms of prognosis, EGFR mutation correlates with poor survival and cancer progression, whereas ERBB2 is suggested to be overexpressed in early tumor stages (96,97). The significant role of EGFR and its mutations for CCA development suggested the employment of Tyrosine Kinase inhibitors (TKi) as a promising therapeutic strategy, similar to what is currently under use in advanced carcinomas (43,76). However, TKi therapy showed only modest benefits in certain CCA patients (98).

Full Table
Ras and Raf are oncogenes and members of the MAPK pathway. Ras mutations have been associated with both intrahepatic and extrahepatic CCA. Indeed, frequent (i.e. G/A transitions in codon 12) and less frequent (i.e. GGT/ GAT and CCA/CAC transitions in the 12th and 61st codons, respectively) Ras point mutations have been reported (80,99,100). On the other hand, mutations of the Raf isoform Braf, contributes with Ras to CCA development. Indeed, no Braf expression was found in human HCC. The most frequent mutation is localized in exons 15 and leads to a T/A change (81).
Beside oncogenes, TSGs are also involved in CCA development and progression. p53, for instance, is involved in protection against aberrant proliferation, including cell cycle arrest and apoptosis (101). p53 inactivation is one of the most common mutations in human cancers and the most frequent among the class of TSGs (102). In CCA, p53 mutations are well-known and many studies have been performed to determine the specific incidence and the type of mutations (85,103) Thus, the aberrant p53 expression was detected by both immunohistochemistry and sequencing studies, as reviewed by Khan et al. (104). p53 mutations occur mainly in exons 5, 6, 7, and 8 as transitions (G:C/A:T) or less commonly as transversion (G-T) (105).
SMAD4 is another TSG that mediates the transforming growth factor (TGF)-β signals (106). The SMAD4/TGF-β signal transduction pathway also negatively regulates epithelial cell growth (107). Loss of SMAD4 activity is a frequent hallmark of gastrointestinal tumors, and has been most frequently observed in CCA arising the distal common bile duct, close to the pancreas, which is, noteworthy, the organ where that mutation occurs more often (86,108,109).
Adenomatous Polyposis Coli (APC) is an additional TSG that regulates different intracellular pathways (110). The typical mechanism of inactivation is characterized by a mutation in one allele followed by loss of heterozygosity (LOH) with complete gene inactivation. The mutation of APC was originally observed in colorectal cancer, but it is currently associated with many other human cancers (111). In CCA cells, APC mutation occurs quite frequently and may be responsible for the early stages of carcinogenesis (87).
Among the allelic losses, lack of 8p22 was found in intrahepatic CCA and may correlate to tumor progression (112).
Epigenetic alterations in CCA
The role of epigenetic alterations in the pathophysiology of CCA is attacking increasing interest (113-115). Although the current knowledge is sparse, the recent technological advances and the attractive possibility to develop novel diagnostic, prognostic and therapeutic options warrant future research. Here, we will provide an overview of the principal and most relevant epigenetic alterations found in CCA.
DNA hypermethylation
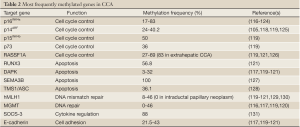
DNA methylation is perhaps the most studied epigenetic change occurring in CCA. The main targets of epigenetic silencing through DNA hypermetilation are TSGs (including those implicated in the regulation of cell cycle and induction of apoptosis), stability genes, and genes involved in inflammatory processes and cell adhesion (Table 2).
Full Table
Among the group of genes involved in the regulation of cell cycle, hypermethylation of p16INK4a is probably the best characterized. p16INK4a, also called cyclin-dependent kinase inhibitor 2A (CDKN2A), binds to cyclin-dependent kinase 4 and inhibits its ability to interact with cyclin D2, thereby preventing the cell to enter in the cell cycle S phase (132). The p16INK4a promoter hypermethylation leads to cell proliferation and oncogenesis. Rates of hypermethylation in the p16INK4a promoter range from 17% to 83% in different studies (116-121). Interestingly, not only p16INK4a hypermethylation seems to be a common event in PSC-related CCA (122) but it has also been associated with a poor clinical outcome (117). Moreover, p16INK4a hypermethylation is thought to be an early event in the progression of CCA: indeed, downregulation of p16INK4a expression was found from intraductal papillary neoplasm of liver and CCA arising from hepatolithiasis (123,124).
Closely related to p16INK4a is p14ARF, the β transcript of the same gene located on chromosome region 9p21. In normal cells, p14ARF blocks the progression from G1 to G2 phase of the cell cycle and inhibits growth of abnormal cells by indirectly p53 activation (133,134). In different studies, the reported methylation frequencies in CCA range from 24% to 40.2%, with the highest value registered in liver fluke-related CCA (105,118,119,125). Interestingly, methylation of p14ARF, DAPK, and/or ASC (see below), together with p53 mutations, were recently reported to correlate with poorly differentiated tumors and poor prognosis (105).
On the same chromosome region 9p21, adjacent to p16INK4a, is located the p15INK4b sequence, which is thought to be an effector of TGF-β-mediated cell cycle arrest (135). Hypermetilation of p15INK4b promoter was reported in 50% of 72 cases of CCA (119). Similarly, 36% methylation of the p73 promoter was also shown in CCA. p73 is a member of the p53 family that is also able to induce cell cycle arrest and apoptosis (136).
RASSF1A, a gene involved in cell cycle regulation, is epigenetically inactivated in CCA. This TSG has been shown to block the cell cycle progression by inhibiting the accumulation of cyclin D1 (137) and the progression of cellular mitosis (138). Hypermethylation of RASSF1A promoter occurs in up to 69% of the patients (126) and, of note, a higher prevalence has been reported in extrahepatic CCA compared with intrahepatic CCA (83% vs. 47%, respectively) (120).
A second subclass of TSGs comprises those involved in promoting apoptosis, the programmed cell death. Hypermethylation in the promoter region of a number of these genes has been found in different studies. Runt-related transcription factor 3 (RUNX3) is a TSG involved in cell growth regulation and TGF-β-induced apoptosis (139). Hypermethylation of RUNX3 promoter was described in up to 56.8% of biliary tract cancers (121). In the same study, the methylation of RUNX3 promoter was more frequent in elderly patients, and was associated with a lower survival rate compared to patients with an unmethylated gene. The methylation of RUNX3 promoter gradually increases from normal samples to biliary intraepithelial neoplasia and eventually CCA (140). In addition, an assay for the analysis of RUNX3, CCND2, CDH13, GRIN2B, and TWIST1 promoter methylation showed increased values in extrahepatic CCAs compared to control tissues (141).
A second member of this subclass of TSGs is the death-associated protein kinase (DAPK). DAPK is a pro-apoptotic mediator of interferon-γ-induced programmed cell death. Hypermethylation of DAPK promoter ranges from 3% to 32% in biliary cancers (117,119-121). Furthermore, it is likely that DAPK methylation correlates with poor prognosis and less survival (105,119,121,142). Additional pro-apoptotic genes found hypermethylated in CCA are semaphorin 3B (SEMA3B) and Target of Methylation-mediated Silencing/Apoptosis Speck like protein containing a CARD (TMS1/ASC). SEMA3B was found to be hypermethylated in 100% of 15 CCA tissue samples (127), while TMS1/ASC showed a 36.1% methylation (128).
Enzymes that participate in DNA repair compose the class of stability genes. Loss-of-function mutations of these genes lead to accumulation of mutations and genomic instability (143). The genes involved in DNA mismatch repair are important for cell protection to possible errors occurring during DNA replication. Defects in DNA mismatch repair machinery have been linked to microsatellite instability (144,145) and demonstrated in a variety of tumors (146,147). Human mutL homologue 1 (hMLH1) is a DNA mismatch repair gene located at 3p21.3 locus. The methylation frequencies of the hMLH1 promoter vary in different studies between 0% in intraductal papillary neoplasms of the biliary tract (129) and 46% in a cohort of 37 patients with biliary tract cancers including gallbladder tumors (120). Interestingly, a high prevalence (62.5%) of microsatellite instability was reported in Thorotrast-induced intrahepatic CCA, suggesting the hypermethylation of the hMLH1 promoter may be in part the cause of this phenomenon (148). Moreover, the same epigenetic process has been reported in 44.6% of cases of liver fluke-related CCA with a significant association with poorly differentiated subtype (130).
An alternative enzyme involved in DNA repair is the O6-methylguanine-DNA methyltransferase (MGMT). The frequency of MGMT promoter methylation seems to vary between different CCA reports, from 33-49% (119,120) to 0% depending on the selected group of patients with CCA (116,117). However, interestingly, the lack of MGMT immunohistochemical staining correlates with poor prognosis of extrahepatic CCA (149).
As mentioned above, chronic biliary inflammation predisposes to the development of CCA (110,150). In this context, IL-6, which is found upregulated in the course of inflammation, is a pivotal growth and survival cytokine in CCA (75) by promoting the expression of the potent anti-apoptotic protein myeloid cell leukemia 1 (MLC1) via phosphorylation of STAT-3 (151). Under physiological conditions, IL-6 induces the expression of the suppressor of cytokine signal 3 (SOCS-3), which in turn inhibits IL-6 signal in a classic feedback loop (152). Interestingly, experimental hypermethylation of SOCS-3 promoter that occurs in a subset of CCAs is responsible for sustained IL-6/STAT-3 signaling and enhanced MLC1 expression (131). These data suggest the use of demethylating agents as a therapeutic approach to revert this process.
Cell adhesion proteins may also be affected by epigenetic silencing in CCA through their gene promoter hypermethylation. Thus, alterations in the expression and function of cadherins, important cell adhesion proteins; are thought to be involved in the epithelial to mesenchymal transition (EMT) (153) and therefore to contribute to tumor progression and metastasis (154,155). The hypermethylation rates of E(epithelial)-cadherin promoter in CCA ranges between 21.5% and 43% (117,119-121). In this regard, a correlation between promoter methylation and reduced protein expression, measured by immunohistochemistry, was reported (117).
Histone modification
To date, limited evidences about the role of histone modifications in CCA exists. However, the intriguing possibility to open novel therapeutic approaches guarantees future research efforts in the upcoming years (156). So far, experimental incubation of different human CCA cell lines with HDAC inhibitors (i.e. MS-275, trichostatin A, NVP-LAQ824, and NVPLBH589) resulted in cell growth arrest and reduced survival in a dose-dependent manner (157-159). Moreover, the combination of conventional cytostatic drugs, such as gemcitabine or doxorubicin, or new agents such as sorafenib or bortezomib, and MS-275 resulted in additive or synergic growth inhibitory effect (157), via induction of apoptosis and cell cycle arrest (157,159). Importantly, initial evidences for the potential therapeutic role of HDAC inhibitors in vivo were reported. Thus, administration of the histone deacetylase inhibitor NVPLBH589 to nude mice with subcutaneously generated CCA tumors significantly reduced the tumor mass and also potentiated the efficacy of gemcitabine (159). Moreover, HDAC1 overexpression correlates with malignant behavior and poor intrahepatic CCA prognosis (160).
MicroRNAs
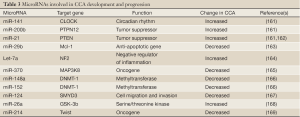
An evident role for microRNAs in CCA biology has been emerging in the last years (Table 3). Previous reports have focused on the study of microRNA expression in different CCA cell lines and shed light, at least in part, on the mechanisms governing their biology and function. A number of microRNAs (e.g., miR-141, miR-200b, miR-21, miR-29b) have been described to be either up or downregulated in CCA cell lines (161,163), and their predicted targets were found to be associated with cell growth and apoptosis.
Full Table
The first microRNA profile comparing human intrahepatic CCA and normal cholangiocyte cell lines was based on cloning methodology and identified eight microRNAs specifically downregulated in cancer cell lines (i.e. miR-22, miR-125a, miR-127, miR-199a, miR-199*, miR-214, miR-376a, and miR-424) (170). In addition, a complex interplay between promoter hypermethylation, inflammation signals and microRNAs expression has been described. Thus, overexpression of IL-6 in human CCA cell lines was shown to increase the levels of microRNA let-7a, which in turn contributes to the survival effect of IL-6 by increasing the phosphorylation of STAT-3 (164). Furthermore, this cytokine increases the expression of the DNA methyltransferase enzyme-1 (DNMT-1) that epigenetically inhibits the transcription of miR-370, resulting in MAP3K8-dependent cell growth (165). Moreover, IL-6 can directly modulate the expression of both miR-148a and miR-152, which in turn regulate the expression of DNMT-1 and TSG (166).
An interesting interplay between epigenetic regulation of microRNAs and Hepatitis C core proteins has also been recently reported (167). The authors showed that downregulation of miR-124—characteristic of HCV-related intrahepatic CCA—is induced in vitro by the HCV core proteins through epigenetic silencing via DNMT-1 upregulation.
SMYD3 was identified as a potential target gene of miR-124 and found to be involved in miR-124 mediated migration and invasion of CCA cells.
The role of microRNAs has been also investigated in human tissue samples. Thus, a genome wide microRNA expression pattern was performed using laser micro dissection techniques comparing 27 intrahepatic CCAs, 10 normal cholangiocyte cell samples, and normal liver tissues. The results showed 38 microRNAs differentially expressed between normal and tumoral samples (171).
miR-21 and miR-26a were found highly overexpressed in CCA. While miR-21 expression was detected with a sensitivity of 95% and 100% of specificity (162) miR-26a was found in 90.5% of the CCA samples and only in 33.3% of controls (168). In addition, miR-26a was shown to promote CCA growth both in vitro and in vivo by direct targeting the levels of glycogen synthase kinase 3β (GSK-3β), which normally regulates the degradation of β-catenin. The subsequent accumulation of β-catenin stimulated the transcription of different genes involved in tumor growth, such as c-Myc, cyclinD1, and peroxisome proliferator-activated receptor δ (168).
On the other hand, microRNAs were indicated to play an important role in the regulation of the metastasis of intrehepatic CCA. Indeed, miR-214 expression was found downregulated in intrahepatic CCAs from patients who developed metastasis compared to non-metastatic CCA tumors (169). The authors showed an indirect correlation between miR-214 levels and Twist, an important inhibitor of E-cadherin transcription, suggesting a potential role of miR-214 regulating the epithelial-mesenchymal transition of the tumor.
Conclusions
CCA is a deadly disease with an incidence increasing worldwide. Although the knowledge on the pathogenesis and the clinical features of the disease has significantly been improved, CCA still represents a major challenge for clinicians. Diagnosis is mostly performed when the disease is already at an advanced stage, thus making the medical and surgical therapy largely ineffective. The successes achieved in the management of different cancers have been commonly based on the identifications of categories of patients at risk and on the consequent set up of surveillance protocols. In this regard, in colon cancer, the identification of familial genetically-based predispositions have led to the determination of specific endoscopic surveillance for offsprings affected patients, increasing the rates of early diagnosis and survival. Identification of patients with high risk for CCA development is the next challenge for the translational research in the upcoming years. In particular, the identification of how genetic and epigenetic modifications may play a major role in CCA development, progression, and metastasis may open a new era for the management of CCA, and may represent a potential strategy for the treatment of this devastating malignancy.
Acknowledgements
This work was supported by a MIUR grant PRIN 2009 - prot. 2009X84L84_003 and Ministero della Salute grant GR-2010-2306996 to Dr. Marzioni.
Disclosure: The authors declare no conflict of interest.
References
- Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology 2005;128:1655-67. [PubMed]
- Cardinale V, Semeraro R, Torrice A, et al. Intra-hepatic and extra-hepatic cholangiocarcinoma: New insight into epidemiology and risk factors. World J Gastrointest Oncol 2010;2:407-16. [PubMed]
- Mosconi S, Beretta GD, Labianca R, et al. Cholangiocarcinoma. Crit Rev Oncol Hematol 2009;69:259-70. [PubMed]
- Khan SA, Thomas HC, Davidson BR, et al. Cholangiocarcinoma. Lancet 2005;366:1303-14. [PubMed]
- Patel AH, Harnois DM, Klee GG, et al. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol 2000;95:204-7. [PubMed]
- Gores GJ. Early detection and treatment of cholangiocarcinoma. Liver Transpl 2000;6:S30-4. [PubMed]
- Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin Liver Dis 2004;24:139-54. [PubMed]
- Patel T. Cholangiocarcinoma--controversies and challenges. Nat Rev Gastroenterol Hepatol 2011;8:189-200. [PubMed]
- Murakami Y, Uemura K, Sudo T, et al. Prognostic factors after surgical resection for intrahepatic, hilar, and distal cholangiocarcinoma. Ann Surg Oncol 2011;18:651-8. [PubMed]
- Akamatsu N, Sugawara Y, Hashimoto D. Surgical strategy for bile duct cancer: Advances and current limitations. World J Clin Oncol 2011;2:94-107. [PubMed]
- Khan SA, Davidson BR, Goldin RD, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 2012;61:1657-69. [PubMed]
- Yoshida T, Matsumoto T, Sasaki A, et al. Prognostic factors after pancreatoduodenectomy with extended lymphadenectomy for distal bile duct cancer. Arch Surg 2002;137:69-73. [PubMed]
- Gatto M, Alvaro D. New insights on cholangiocarcinoma. World J Gastrointest Oncol 2010;2:136-45. [PubMed]
- Vauthey JN, Blumgart LH. Recent advances in the management of cholangiocarcinomas. Semin Liver Dis 1994;14:109-14. [PubMed]
- Khan SA, Toledano MB, Taylor-Robinson SD. Epidemiology, risk factors, and pathogenesis of cholangiocarcinoma. HPB (Oxford) 2008;10:77-82. [PubMed]
- Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology 2001;33:1353-7. [PubMed]
- Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004;24:115-25. [PubMed]
- Taylor-Robinson SD, Toledano MB, Arora S, et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968-1998. Gut 2001;48:816-20. [PubMed]
- West J, Wood H, Logan RF, et al. Trends in the incidence of primary liver and biliary tract cancers in England and Wales 1971-2001. Br J Cancer 2006;94:1751-8. [PubMed]
- von Hahn T, Ciesek S, Wegener G, et al. Epidemiological trends in incidence and mortality of hepatobiliary cancers in Germany. Scand J Gastroenterol 2011;46:1092-8. [PubMed]
- Alvaro D, Crocetti E, Ferretti S, et al. Descriptive epidemiology of cholangiocarcinoma in Italy. Dig Liver Dis 2010;42:490-5. [PubMed]
- Jepsen P, Vilstrup H, Tarone RE, et al. Incidence rates of intra- and extrahepatic cholangiocarcinomas in Denmark from 1978 through 2002. J Natl Cancer Inst 2007;99:895-7. [PubMed]
- Lepage C, Cottet V, Chauvenet M, et al. Trends in the incidence and management of biliary tract cancer: a French population-based study. J Hepatol 2011;54:306-10. [PubMed]
- Welzel TM, McGlynn KA, Hsing AW, et al. Impact of classification of hilar cholangiocarcinomas (Klatskin tumors) on the incidence of intra- and extrahepatic cholangiocarcinoma in the United States. J Natl Cancer Inst 2006;98:873-5. [PubMed]
- Khan SA, Emadossadaty S, Ladep NG, et al. Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J Hepatol 2012;56:848-54. [PubMed]
- McLean L, Patel T. Racial and ethnic variations in the epidemiology of intrahepatic cholangiocarcinoma in the United States. Liver Int 2006;26:1047-53. [PubMed]
- Sripa B, Pairojkul C. Cholangiocarcinoma: lessons from Thailand. Curr Opin Gastroenterol 2008;24:349-56. [PubMed]
- Sripa B, Kaewkes S, Sithithaworn P, et al. Liver fluke induces cholangiocarcinoma. PLoS Med 2007;4:e201. [PubMed]
- Songserm N, Promthet S, Sithithaworn P, et al. MTHFR polymorphisms and Opisthorchis viverrini infection: a relationship with increased susceptibility to cholangiocarcinoma in Thailand. Asian Pac J Cancer Prev 2011;12:1341-5. [PubMed]
- Honjo S, Srivatanakul P, Sriplung H, et al. Genetic and environmental determinants of risk for cholangiocarcinoma via Opisthorchis viverrini in a densely infested area in Nakhon Phanom, northeast Thailand. Int J Cancer 2005;117:854-60. [PubMed]
- Yuspa SH. Overview of carcinogenesis: past, present and future. Carcinogenesis 2000;21:341-4. [PubMed]
- Russo A, Zanna I, Tubiolo C, et al. Hereditary common cancers: molecular and clinical genetics. Anticancer Res 2000;20:4841-51. [PubMed]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24. [PubMed]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004;10:789-99. [PubMed]
- Croce CM. Molecular origins of cancer: Oncogenes and cancer. N Engl J Med 2008;358:502-11. [PubMed]
- Loeb KR, Loeb LA. Significance of multiple mutations in cancer. Carcinogenesis 2000;21:379-85. [PubMed]
- Wallis YL, Macdonald F. Demystified .. oncogenes. Mol Pathol 1999;52:55-63. [PubMed]
- Mathas S, Hinz M, Anagnostopoulos I, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J 2002;21:4104-13. [PubMed]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [PubMed]
- Liu X, Newton RC, Scherle PA. Developing c-MET pathway inhibitors for cancer therapy: progress and challenges. Trends Mol Med 2010;16:37-45. [PubMed]
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 2009;21:177-84. [PubMed]
- Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys 2004;59:21-6. [PubMed]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature Reviews. Cancer 2005;5:341-54. [PubMed]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989;49:4682-9. [PubMed]
- Schuetz JM, Johnson NA, Morin RD, et al. BCL2 mutations in diffuse large B-cell lymphoma. Leukemia 2012;26:1383-90. [PubMed]
- Sherr CJ. Principles of tumor suppression. Cell 2004;116:235-46. [PubMed]
- Hodgson S. Mechanisms of inherited cancer susceptibility. J Zhejiang Univ Sci B 2008;9:1-4. [PubMed]
- Suzuki K, Matsubara H. Recent advances in p53 research and cancer treatment. J Biomed Biotechnol 2011;2011:978312.
- Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol 2010;20:281-93. [PubMed]
- Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res 2010;70:431-5. [PubMed]
- Hassen S, Ali N, Chowdhury P. Molecular signaling mechanisms of apoptosis in hereditary non-polyposis colorectal cancer. World J Gastrointest Pathophysiol 2012;3:71-9. [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [PubMed]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915-26. [PubMed]
- Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol 2002;12:389-98. [PubMed]
- Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet 2005;6:403-10. [PubMed]
- Bestor TH. Transposons reanimated in mice. Cell 2005;122:322-5. [PubMed]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6-21. [PubMed]
- Bird A. The essentials of DNA methylation. Cell 1992;70:5-8. [PubMed]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000;16:168-74. [PubMed]
- Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A 1994;91:9700-4. [PubMed]
- Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995;55:4525-30. [PubMed]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301:89-92. [PubMed]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107-16. [PubMed]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature 1992;355:219-24. [PubMed]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem 2001;70:81-120. [PubMed]
- Thiagalingam S, Cheng KH, Lee HJ, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci 2003;983:84-100. [PubMed]
- Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol 2009;21:416-25. [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [PubMed]
- Knowling S, Morris KV. Non-coding RNA and antisense RNA. Nature's trash or treasure? Biochimie 2011;93:1922-7. [PubMed]
- Manikandan J, Aarthi JJ, Kumar SD, et al. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation 2008;2:330-4. [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [PubMed]
- Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer 2010;46:298-311. [PubMed]
- Gores GJ. Cholangiocarcinoma: current concepts and insights. Hepatology 2003;37:961-9. [PubMed]
- Wehbe H, Henson R, Meng F, et al. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res 2006;66:10517-24. [PubMed]
- Johnson C, Han Y, Hughart N, et al. Interleukin-6 and its receptor, key players in hepatobiliary inflammation and cancer. Transl Gastrointest Cancer 2012;1:58-70. [PubMed]
- Sirica AE. Cholangiocarcinoma: molecular targeting strategies for chemoprevention and therapy. Hepatology 2005;41:5-15. [PubMed]
- Jaiswal M, LaRusso NF, Burgart LJ, et al. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res 2000;60:184-90. [PubMed]
- Jaiswal M, LaRusso NF, Shapiro RA, et al. Nitric oxide-mediated inhibition of DNA repair potentiates oxidative DNA damage in cholangiocytes. Gastroenterology 2001;120:190-9. [PubMed]
- Nzeako UC, Guicciardi ME, Yoon JH, et al. COX-2 inhibits Fas-mediated apoptosis in cholangiocarcinoma cells. Hepatology 2002;35:552-9. [PubMed]
- O’Dell MR, Huang JL, Whitney-Miller CL, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res 2012;72:1557-67. [PubMed]
- Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut 2003;52:706-12. [PubMed]
- Leone F, Cavalloni G, Pignochino Y, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clin Cancer Res 2006;12:1680-5. [PubMed]
- Kiguchi K, Carbajal S, Chan K, et al. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res 2001;61:6971-6. [PubMed]
- Socoteanu MP, Mott F, Alpini G, et al. c-Met targeted therapy of cholangiocarcinoma. World J Gastroenterol 2008;14:2990-4. [PubMed]
- Arora DS, Ramsdale J, Lodge JP, et al. p53 but not bcl-2 is expressed by most cholangiocarcinomas: a study of 28 cases. Histopathology 1999;34:497-501. [PubMed]
- Kang YK, Kim WH, Jang JJ. Expression of G1-S modulators (p53, p16, p27, cyclin D1, Rb) and Smad4/Dpc4 in intrahepatic cholangiocarcinoma. Hum Pathol 2002;33:877-83. [PubMed]
- Cong WM, Bakker A, Swalsky PA, et al. Multiple genetic alterations involved in the tumorigenesis of human cholangiocarcinoma: a molecular genetic and clinicopathological study. J Cancer Res Clin Oncol 2001;127:187-92. [PubMed]
- Hassid VJ, Orlando FA, Awad ZT, et al. Genetic and molecular abnormalities in cholangiocarcinogenesis. Anticancer Res 2009;29:1151-6. [PubMed]
- Terada T, Nakanuma Y, Sirica AE. Immunohistochemical demonstration of MET overexpression in human intrahepatic cholangiocarcinoma and in hepatolithiasis. Hum Pathol 1998;29:175-80. [PubMed]
- Miyamoto M, Ojima H, Iwasaki M, et al. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Brit J Cancer 2011;105:131-8. [PubMed]
- Wosikowski K, Schuurhuis D, Johnson K, et al. Identification of epidermal growth factor receptor and c-erbB2 pathway inhibitors by correlation with gene expression patterns. J Natl Cancer Inst 1997;89:1505-15. [PubMed]
- Ito Y, Takeda T, Sasaki Y, et al. Expression and clinical significance of the erbB family in intrahepatic cholangiocellular carcinoma. Pathol Res Pract 2001;197:95-100. [PubMed]
- Ukita Y, Kato M, Terada T. Gene amplification and mRNA and protein overexpression of c-erbB-2 (HER-2/neu) in human intrahepatic cholangiocarcinoma as detected by fluorescence in situ hybridization, in situ hybridization, and immunohistochemistry. J Hepatol 2002;36:780-5. [PubMed]
- Sirica AE, Lai GH, Endo K, et al. Cyclooxygenase-2 and ERBB-2 in cholangiocarcinoma: potential therapeutic targets. Semin Liver Dis 2002;22:303-13. [PubMed]
- Lai GH, Zhang Z, Shen XN, et al. erbB-2/neu transformed rat cholangiocytes recapitulate key cellular and molecular features of human bile duct cancer. Gastroenterology 2005;129:2047-57. [PubMed]
- Yoshikawa D, Ojima H, Iwasaki M, et al. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br J Cancer 2008;98:418-25. [PubMed]
- Harder J, Waiz O, Otto F, et al. EGFR and HER2 expression in advanced biliary tract cancer. World J Gastroenterol 2009;15:4511-7. [PubMed]
- Andersen JB, Spee B, Blechacz BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012;142:1021-31. [PubMed]
- Levi S, Urbano-Ispizua A, Gill R, et al. Multiple K-ras codon 12 mutations in cholangiocarcinomas demonstrated with a sensitive polymerase chain reaction technique. Cancer Res 1991;51:3497-502. [PubMed]
- Ohashi K, Tstsumi M, Nakajima Y, et al. Ki-ras point mutations and proliferation activity in biliary tract carcinomas. Br J Cancer 1996;74:930-5. [PubMed]
- Jin S, Levine AJ. The p53 functional circuit. J Cell Sci 2001;114:4139-40. [PubMed]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012;26:1268-86. [PubMed]
- Washington K, Gottfried MR. Expression of p53 in adenocarcinoma of the gallbladder and bile ducts. Liver 1996;16:99-104. [PubMed]
- Khan SA, Thomas HC, Toledano MB, et al. p53 Mutations in human cholangiocarcinoma: a review. Liver Int 2005;25:704-16. [PubMed]
- Xiaofang L, Kun T, Shaoping Y, et al. Correlation between promoter methylation of p14(ARF), TMS1/ASC, and DAPK, and p53 mutation with prognosis in cholangiocarcinoma. World J Surg Oncol 2012;10:5. [PubMed]
- Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res 2012;347:21-36. [PubMed]
- Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun 2003;306:799-804. [PubMed]
- Argani P, Shaukat A, Kaushal M, et al. Differing rates of loss of DPC4 expression and of p53 overexpression among carcinomas of the proximal and distal bile ducts. Cancer 2001;91:1332-41. [PubMed]
- Rijken AM, Hu J, Perlman EJ, et al. Genomic alterations in distal bile duct carcinoma by comparative genomic hybridization and karyotype analysis. Genes Chromosomes Cancer 1999;26:185-91. [PubMed]
- Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta 1997;1332:F127-47. [PubMed]
- Ichihashi N, Kitajima Y. Loss of heterozygosity of adenomatous polyposis coli gene in cutaneous tumors as determined by using polymerase chain reaction and paraffin section preparations. J Dermatol Sci 2000;22:102-6. [PubMed]
- Kawaki J, Miyazaki M, Ito H, et al. Allelic loss in human intrahepatic cholangiocarcinoma: correlation between chromosome 8p22 and tumor progression. Int J Cancer 2000;88:228-31. [PubMed]
- Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol 2012;28:266-72. [PubMed]
- Isomoto H. Epigenetic alterations associated with cholangiocarcinoma Oncol Rep 2009;22:227-32. [PubMed]
- Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver Int 2008;28:12-27. [PubMed]
- Kim BH, Cho NY, Choi M, et al. Methylation profiles of multiple CpG island loci in extrahepatic cholangiocarcinoma versus those of intrahepatic cholangiocarcinomas. Arch Pathol Lab Med 2007;131:923-30. [PubMed]
- Lee S, Kim WH, Jung HY, et al. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol 2002;161:1015-22. [PubMed]
- Tannapfel A, Sommerer F, Benicke M, et al. Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J Pathol 2002;197:624-31. [PubMed]
- Yang B, House MG, Guo M, et al. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol 2005;18:412-20. [PubMed]
- Koga Y, Kitajima Y, Miyoshi A, et al. Tumor progression through epigenetic gene silencing of O(6)-methylguanine-DNA methyltransferase in human biliary tract cancers. Ann Surg Oncol 2005;12:354-63. [PubMed]
- Tozawa T, Tamura G, Honda T, et al. Promoter hypermethylation of DAP-kinase is associated with poor survival in primary biliary tract carcinoma patients. Cancer Sci 2004;95:736-40. [PubMed]
- Ahrendt SA, Eisenberger CF, Yip L, et al. Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. J Surg Res 1999;84:88-93. [PubMed]
- Ishikawa A, Sasaki M, Sato Y, et al. Frequent p16ink4a inactivation is an early and frequent event of intraductal papillary neoplasm of the liver arising in hepatolithiasis. Hum Pathol 2004;35:1505-14. [PubMed]
- Sasaki M, Yamaguchi J, Itatsu K, et al. Over-expression of polycomb group protein EZH2 relates to decreased expression of p16 INK4a in cholangiocarcinogenesis in hepatolithiasis. J Pathol 2008;215:175-83. [PubMed]
- Chinnasri P, Pairojkul C, Jearanaikoon P, et al. Preferentially different mechanisms of inactivation of 9p21 gene cluster in liver fluke-related cholangiocarcinoma. Hum Pathol 2009;40:817-26. [PubMed]
- Wong N, Li L, Tsang K, et al. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol 2002;37:633-9. [PubMed]
- Tischoff I, Markwarth A, Witzigmann H, et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer 2005;115:684-9. [PubMed]
- Liu XF, Zhu SG, Zhang H, et al. The methylation status of the TMS1/ASC gene in cholangiocarcinoma and its clinical significance. Hepatobiliary Pancreat Dis Int 2006;5:449-53. [PubMed]
- Abraham SC, Lee JH, Boitnott JK, et al. Microsatellite instability in intraductal papillary neoplasms of the biliary tract. Mod Pathol 2002;15:1309-17. [PubMed]
- Limpaiboon T, Khaenam P, Chinnasri P, et al. Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma. Cancer Lett 2005;217:213-9. [PubMed]
- Isomoto H, Mott JL, Kobayashi S, et al. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology 2007;132:384-96. [PubMed]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993;366:704-7. [PubMed]
- Silva J, Silva JM, Dominguez G, et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol 2003;199:289-97. [PubMed]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998;92:725-34. [PubMed]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 1994;371:257-61. [PubMed]
- Maas AM, Bretz AC, Mack E, et al. Targeting p73 in cancer. Cancer Lett 2013;332:229-36. [PubMed]
- Shivakumar L, Minna J, Sakamaki T, et al. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol 2002;22:4309-18. [PubMed]
- Song MS, Song SJ, Ayad NG, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nature Cell Biol 2004;6:129-37. [PubMed]
- Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene 2004;23:4336-40. [PubMed]
- Kim BH, Cho NY, Shin SH, et al. CpG island hypermethylation and repetitive DNA hypomethylation in premalignant lesion of extrahepatic cholangiocarcinoma. Virchows Archiv 2009;455:343-51. [PubMed]
- Shin SH, Lee K, Kim BH, et al. Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity. J Mol Diagn 2012;14:256-63. [PubMed]
- Liu XF, Kong FM, Xu Z, et al. Promoter hypermethylation of death-associated protein kinase gene in cholangiocarcinoma. Hepatobiliary Pancreat Dis Int 2007;6:407-11. [PubMed]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287-94. [PubMed]
- Fleisher AS, Esteller M, Tamura G, et al. Hypermethylation of the hMLH1 gene promoter is associated with microsatellite instability in early human gastric neoplasia. Oncogene 2001;20:329-35. [PubMed]
- Thibodeau SN, French AJ, Roche PC, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 1996;56:4836-40. [PubMed]
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998;95:6870-5. [PubMed]
- Esteller M, Levine R, Baylin SB, et al. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene 1998;17:2413-7. [PubMed]
- Liu D, Momoi H, Li L, et al. Microsatellite instability in thorotrast-induced human intrahepatic cholangiocarcinoma. Int J Cancer 2002;102:366-71. [PubMed]
- Kohya N, Miyazaki K, Matsukura S, et al. Deficient expression of O(6)-methylguanine-DNA methyltransferase combined with mismatch-repair proteins hMLH1 and hMSH2 is related to poor prognosis in human biliary tract carcinoma. Ann Surg Oncol 2002;9:371-9. [PubMed]
- Berthiaume EP, Wands J. The molecular pathogenesis of cholangiocarcinoma. Semin Liver Dis 2004;24:127-37. [PubMed]
- Isomoto H, Kobayashi S, Werneburg NW, et al. Interleukin 6 upregulates myeloid cell leukemia-1 expression through a STAT3 pathway in cholangiocarcinoma cells. Hepatology 2005;42:1329-38. [PubMed]
- Croker BA, Krebs DL, Zhang JG, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol 2003;4:540-5. [PubMed]
- Chua HL, Bhat-Nakshatri P, Clare SE, et al. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 2007;26:711-24. [PubMed]
- Stemmler MP. Cadherins in development and cancer. Mol Biosyst 2008;4:835-50. [PubMed]
- Makrilia N, Kollias A, Manolopoulos L, et al. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest 2009;27:1023-37. [PubMed]
- Kouraklis G, Theocharis S. Histone deacetylase inhibitors: a novel target of anticancer therapy Oncol Rep 2006;15:489-94. [PubMed]
- Baradari V, Hopfner M, Huether A, et al. Histone deacetylase inhibitor MS-275 alone or combined with bortezomib or sorafenib exhibits strong antiproliferative action in human cholangiocarcinoma cells. World J Gastroenterol 2007;13:4458-66. [PubMed]
- Xu LN, Wang X, Zou SQ. Effect of histone deacetylase inhibitor on proliferation of biliary tract cancer cell lines. World J Gastroenterol 2008;14:2578-81. [PubMed]
- Bluethner T, Niederhagen M, Caca K, et al. Inhibition of histone deacetylase for the treatment of biliary tract cancer: a new effective pharmacological approach. World J Gastroenterol 2007;13:4761-70. [PubMed]
- Morine Y, Shimada M, Iwahashi S, et al. Role of histone deacetylase expression in intrahepatic cholangiocarcinoma. Surgery 2012;151:412-9. [PubMed]
- Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006;130:2113-29. [PubMed]
- Selaru FM, Olaru AV, Kan T, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 2009;49:1595-601. [PubMed]
- Mott JL, Kobayashi S, Bronk SF, et al. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 2007;26:6133-40. [PubMed]
- Meng F, Henson R, Wehbe-Janek H, et al. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem 2007;282:8256-64. [PubMed]
- Meng F, Wehbe-Janek H, Henson R, et al. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 2008;27:378-86. [PubMed]
- Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010;51:881-90. [PubMed]
- Zeng B, Li Z, Chen R, et al. Epigenetic regulation of miR-124 by Hepatitis C Virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett 2012;586:3271-8. [PubMed]
- Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology 2012;143:246-56.e8.
- Li B, Han Q, Zhu Y, et al. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J 2012;279:2393-8. [PubMed]
- Kawahigashi Y, Mishima T, Mizuguchi Y, et al. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J Nippon Med Sch 2009;76:188-97. [PubMed]
- Chen L, Yan HX, Yang W, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol 2009;50:358-69. [PubMed]


